Search Count: 188
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2024-11-20 Classification: HYDROLASE/INHIBITOR Ligands: DMS, EDO, SO4, A1A21, A1AFD, GOL |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2024-11-20 Classification: HYDROLASE/INHIBITOR Ligands: A1AFE, DMS |
 |
Structure Of M66I Mutant Of Disulfide Stabilized Hiv-1 Ca Hexamer In Complex With Cpsf6 Peptide And Ip6
Organism: Human immunodeficiency virus 1, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2024-03-13 Classification: VIRAL PROTEIN Ligands: IHP |
 |
Crystal Structure Of C-Terminal Domain Of Msk1 In Complex With In Covalently Bound Literature Rsk2 Inhibitor Pyrrolopyrimidine Cyanoacrylamide Compound 25 (Co-Crystal)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2022-08-31 Classification: TRANSFERASE Ligands: SUU, OXM |
 |
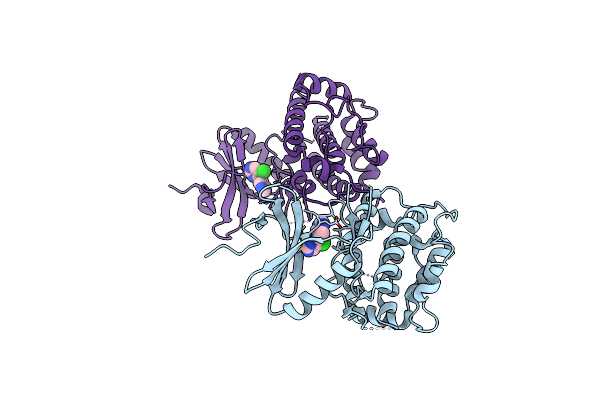
Crystal Structure Of C-Terminal Domain Of Msk1 In Complex With Covalently Bound With Literature Rsk2 Inhibitor Indazole Cyanoacrylamide Compound 26 (Soak)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2022-07-20 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 1LE |
 |
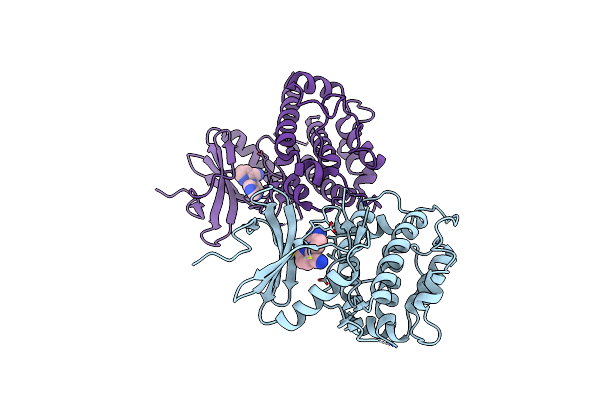
Crystal Structure Of C-Terminal Domain Of Msk1 In Complex With Covalently Bound Pyrrolopyrimidine Compound 20 (Co-Crystal)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2022-07-06 Classification: TRANSFERASE Ligands: O1K |
 |
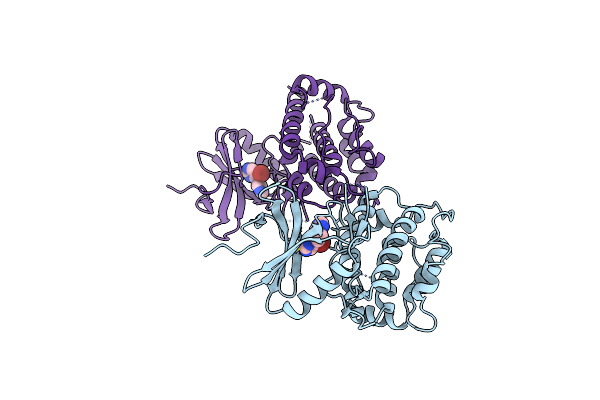
Crystal Structure Of C-Terminal Domain Of Msk1 In Complex With Covalently Bound Pyrrolopyrimidine Compound 23 (Co-Crystal)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2022-07-06 Classification: TRANSFERASE Ligands: O1R, IOD |
 |
Crystal Structure Of C-Terminal Domain Of Msk1 In Complex With Covalently Bound Pyrrolopyrimidine Compound 27 (Co-Crystal)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2022-07-06 Classification: TRANSFERASE Ligands: O10 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.58 Å Release Date: 2019-06-26 Classification: CHAPERONE |
 |
Crystal Structure Of A Hypothetical Protein Rv3716C From Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-05-16 Classification: DNA BINDING PROTEIN Ligands: CD, EDO |
 |
Crystal Structure Of Aspartate-Semialdehyde Dehydrogenase From Neisseria Gonorrhoeae
Organism: Neisseria gonorrhoeae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-10-25 Classification: OXIDOREDUCTASE Ligands: SO4, EDO |
 |
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2017-03-22 Classification: MOTOR PROTEIN Ligands: SO4 |
 |
Crystal Structure Of Polymerase Acid Protein (Pa) From Influenza A Virus, Wilson-Smith/1933 (H1N1) Bound To Ebsi-39 (2,3-Dichlorophenyl)Methanol
Organism: Influenza a virus (strain a/wilson-smith/1933 h1n1)
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2017-03-08 Classification: VIRAL PROTEIN Ligands: DMS, EDO, 6B0 |
 |
Crystal Structure Of Polymerase Acid Protein (Pa) From Influenza A Virus, Wilson-Smith/1933 (H1N1) Bound To Fragment Hit Ebsi-747 1-(4-Chlorophenyl)-1H-Imidazole
Organism: Influenza a virus (strain a/wilson-smith/1933 h1n1)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2017-02-22 Classification: VIRAL PROTEIN Ligands: 1CI, DMS |
 |
Crystal Structure Of Polymerase Acid Protein (Pa) From Influenza A Virus, Wilson-Smith/1933 (H1N1) Bound To Fragment Hit Ebsi-576 (5,6-Dichloro-1H-1,3-Benzodiazol-2-Yl)Methanol
Organism: Influenza a virus (strain a/wilson-smith/1933 h1n1)
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2017-02-22 Classification: VIRAL PROTEIN Ligands: 6B1, DMS, CL |
 |
Crystal Structure Of Polymerase Acid Protein (Pa) From Influenza A Virus, Wilson-Smith/1933 (H1N1) Bound To Fragment Hit Ebsi-279 N-[(4-Chlorophenyl)Methyl]-1-Methyl-1H-Pyrazolo[3,4-D]Pyrimidin-4-Amine
Organism: Influenza a virus (strain a/wilson-smith/1933 h1n1)
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2017-02-22 Classification: VIRAL PROTEIN Ligands: 6AY, DMS |
 |
Crystal Structure Of Polymerase Acid Protein (Pa) From Influenza A Virus, Wilson-Smith/1933 (H1N1) Bound To Fragment Hit Ebsi-2643 5-[(4-Chlorophenyl)Methyl]-1,3,4-Oxadiazol-2-Amine
Organism: Influenza a virus (strain a/wilson-smith/1933 h1n1)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-02-22 Classification: VIRAL PROTEIN Ligands: 6AX, DMS, CL, EDO, GOL |
 |
Crystal Structure Of Polymerase Acid Protein (Pa) From Influenza A Virus, Wilson-Smith/1933 (H1N1) Bound To Follow On Fragment Ebsi-4719 5-Chloro-2-(1H-Imidazol-1-Yl)Aniline
Organism: Influenza a virus (strain a/wilson-smith/1933 h1n1)
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2017-02-22 Classification: VIRAL PROTEIN Ligands: 6AV, DMS, CL |
 |
Crystal Structure Of Polymerase Acid Protein (Pa) From Influenza A Virus, Wilson-Smith/1933 (H1N1) Bound To Follow On Fragment Ebsi-4720 1-(4-Bromophenyl)-1H-Imidazole
Organism: Influenza a virus (strain a/wilson-smith/1933 h1n1)
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2017-02-22 Classification: VIRAL PROTEIN Ligands: 6AR, DMS |
 |
Crystal Structure Of Polymerase Acid Protein (Pa) From Influenza A Virus, Wilson-Smith/1933 (H1N1) Bound To Follow On Fragment Ebsi-4721 1-(4-Fluorophenyl)-1H-Imidazole
Organism: Influenza a virus (strain a/wilson-smith/1933 h1n1)
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2017-02-22 Classification: VIRAL PROTEIN Ligands: 6AQ, DMS |

