Search Count: 77
 |
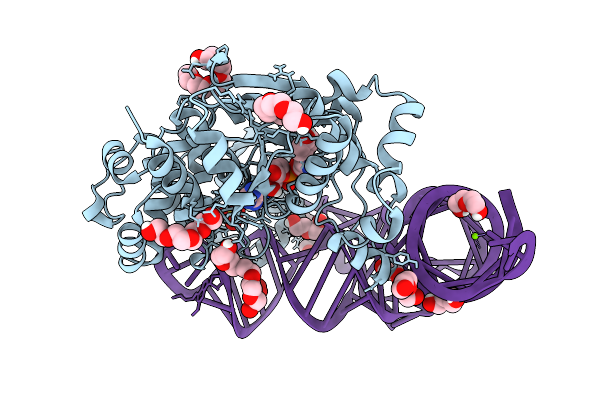
Structure At 1.9 A Resolution Of Thermus Thermophilus Tyrosyl-Trna Synthetase Bound To Wild-Type Trnatyr(Guc).
Organism: Thermus thermophilus hb27, Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2025-09-24 Classification: RNA BINDING PROTEIN Ligands: SO4, PEG, 1PE, MG |
 |
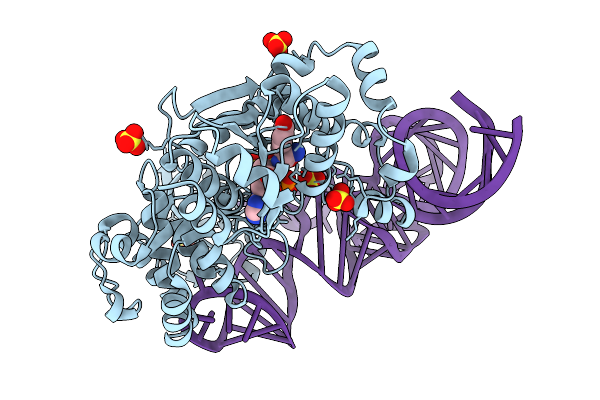
Structure At 1.9 A Resolution Of Thermus Thermophilus Tyrosyl-Trna Synthetase Bound To Wild-Type Modified Trna(Tyr), Tyrosine And Amp
Organism: Thermus thermophilus hb27
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-09-03 Classification: RNA BINDING PROTEIN Ligands: AMP, TYR, PEG, SO4, 1PE, PGE, MG |
 |
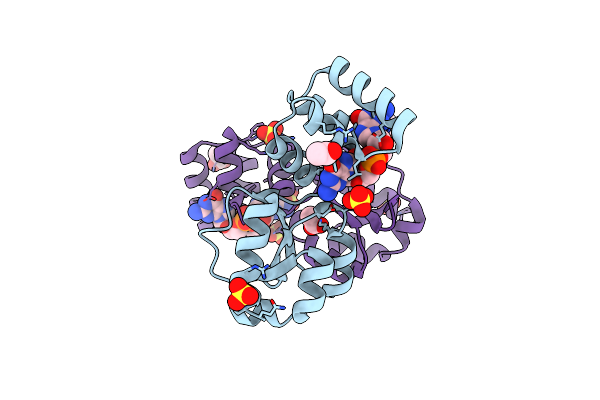
Structure At 2.7 A Resolution Of Thermus Thermophilus Tyrosyl-Trna Synthetase Bound To A Trna(Tyr)(Gua) Transcript, The Sulphamoyl Analogue Of Tyrosyl-Adenylate And Pyrophosphate.
Organism: Thermus thermophilus hb27, Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2025-09-03 Classification: RNA BINDING PROTEIN Ligands: PPV, SO4, YSA |
 |
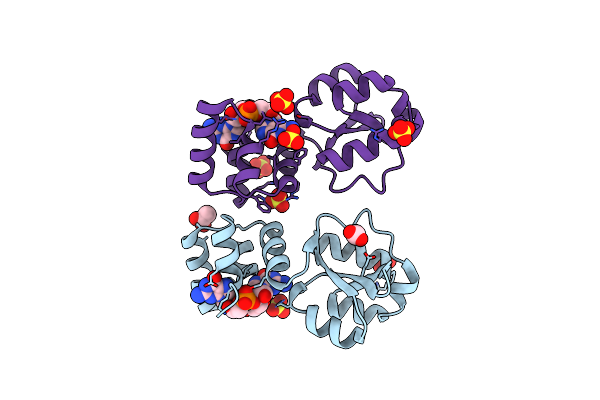
X-Ray Structure Of The Thermus Thermophilus Q190E Mutant Of The Pilf-Gspiib Domain In The C-Di-Gmp Bound State
Organism: Thermus thermophilus hb27
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-06-25 Classification: MOTOR PROTEIN Ligands: C2E, SO4, ACY |
 |
X-Ray Structure Of The Thermus Thermophilus Q218E Mutant Of The Pilf-Gspiib Domain In The C-Di-Gmp Bound State
Organism: Thermus thermophilus hb27
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2025-06-25 Classification: MOTOR PROTEIN Ligands: C2E, SO4, ACT |
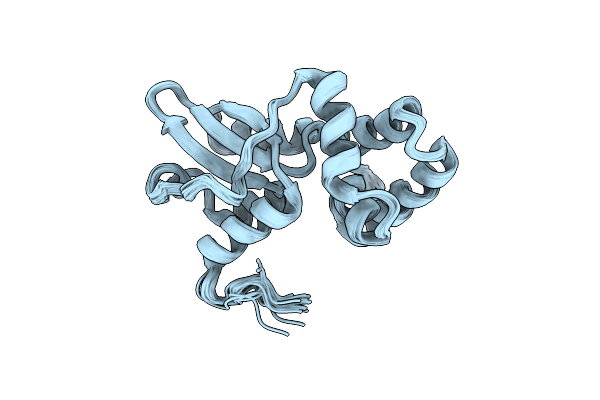 |
Organism: Thermus thermophilus hb27
Method: SOLUTION NMR Release Date: 2025-05-21 Classification: MOTOR PROTEIN |
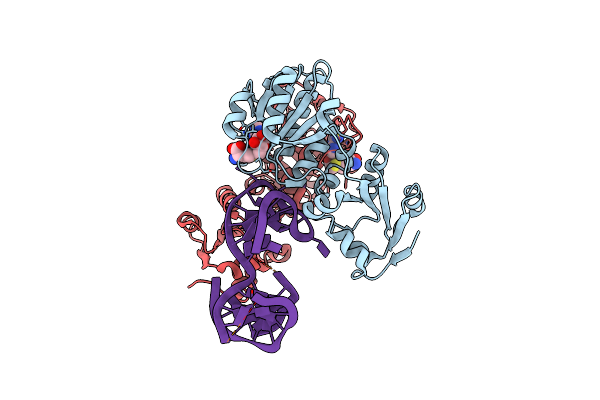 |
Rlmr 23S Rrna Methyltransferase From Thermus Thermophilus In Complex With Rrna And S-Adenosyl-L-Homocysteine (Sah)
Organism: Thermus thermophilus hb27
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2025-04-23 Classification: TRANSFERASE Ligands: SAH |
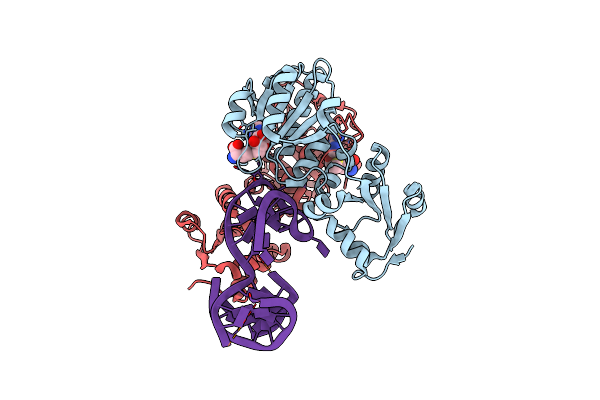 |
Rlmr 23S Rrna Methyltransferase From Thermus Thermophilus In Complex With Methylated Rrna (Um2552) And S-Adenosyl-L-Homocysteine (Sah)
Organism: Thermus thermophilus hb27
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2025-04-23 Classification: TRANSFERASE/RNA Ligands: SAH, SAM |
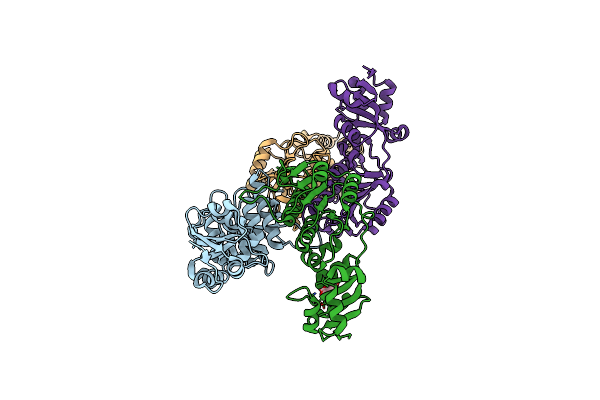 |
Organism: Thermus thermophilus hb27
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2025-04-23 Classification: TRANSFERASE Ligands: CIT |
 |
Crystal Structure Of 3-Hydroxybutyryl-Coa Dehydrogenase From Thermus Thermophilus Hb27 Complexed To Nad+
Organism: Thermus thermophilus hb27
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-03-19 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
Thermus Thermophilus Hb27 Laccase (Tth-Lac) Mutant With Partial Deletion Of Beta-Hairpin Sequence
Organism: Thermus thermophilus hb27
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2025-02-05 Classification: METAL BINDING PROTEIN Ligands: CU, GOL |
 |
Organism: Thermus thermophilus hb27
Method: ELECTRON MICROSCOPY Release Date: 2024-10-16 Classification: PROTEIN FIBRIL Ligands: MG |
 |
Nmr Structure Of The Thermus Thermophilus Pilf-Gspiib Domain In The Apo State
Organism: Thermus thermophilus hb27
Method: SOLUTION NMR Release Date: 2024-07-24 Classification: MOTOR PROTEIN |
 |
Organism: Thermus thermophilus hb27
Method: SOLUTION NMR Release Date: 2024-07-10 Classification: MOTOR PROTEIN Ligands: 5GP |
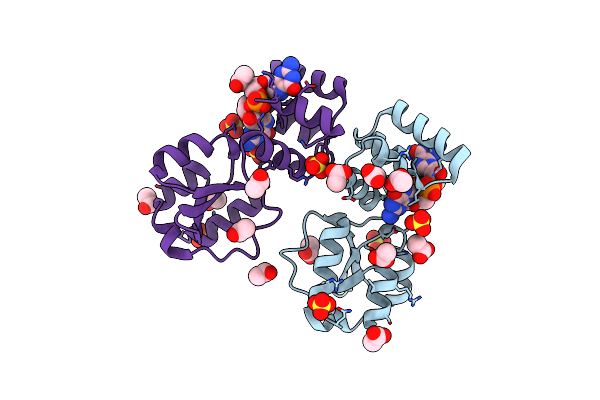 |
X-Ray Structure Of The Thermus Thermophilus Pilf-Gspiib Domain In The C-Di-Gmp Bound State
Organism: Thermus thermophilus hb27
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-06-26 Classification: MOTOR PROTEIN Ligands: C2E, SO4, ACT |
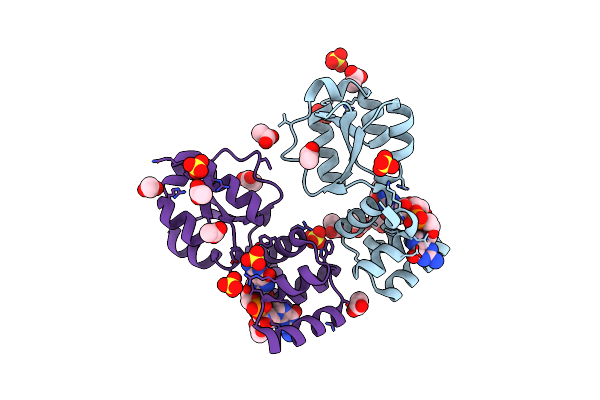 |
X-Ray Structure Of The Thermus Thermophilus K167L Mutant Of The Pilf-Gspiib Domain In The C-Di-Gmp Bound State
Organism: Thermus thermophilus hb27
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-06-26 Classification: MOTOR PROTEIN Ligands: C2E, SO4, ACT |
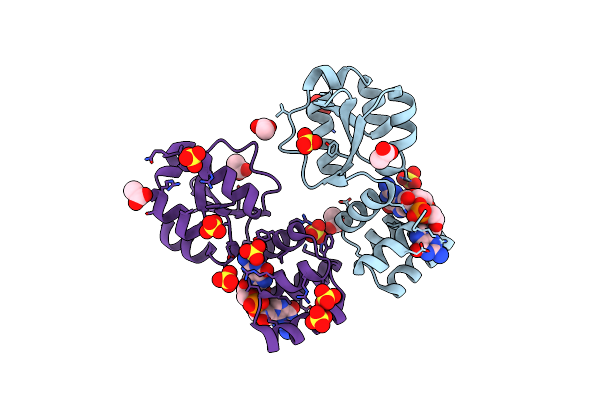 |
X-Ray Structure Of The Thermus Thermophilus K167R Mutant Of The Pilf-Gspiib Domain In The C-Di-Gmp Bound State
Organism: Thermus thermophilus hb27
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-06-26 Classification: MOTOR PROTEIN Ligands: C2E, SO4, ACT |
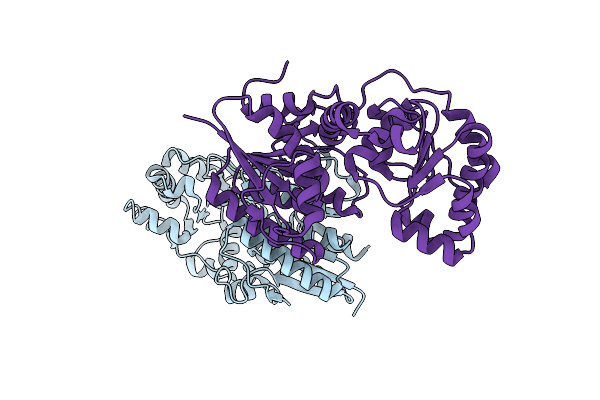 |
X-Ray Crystal Structure Of Udp- 2,3-Diacetamido-2,3-Dideoxy-Glucuronic Acid-2-Epimerase From Thermus Thermophilus Strain Hb27, Apo Form, Ph 9
Organism: Thermus thermophilus hb27
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-09-13 Classification: ISOMERASE Ligands: CL |
 |
X-Ray Crystal Structure Of Udp- 2,3-Diacetamido-2,3-Dideoxy-Glucuronic Acid-2-Epimerase From Thermus Thermophilus Strain Hb27, D98N Mutation, Apo Structure At Ph 6
Organism: Thermus thermophilus hb27
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-09-13 Classification: ISOMERASE Ligands: CL, NA |
 |
X-Ray Crystal Structure Of Udp- 2,3-Diacetamido-2,3-Dideoxy-Glucuronic Acid-2-Epimerase From Thermus Thermophilus Strain Hb27 In Complex With Its Product Udp-2,3-Diacetamido-2,3-Dideoxy-D-Mannuronic Acid At Ph 5
Organism: Thermus thermophilus hb27
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-09-13 Classification: ISOMERASE Ligands: MJ3, CL |

