Search Count: 69
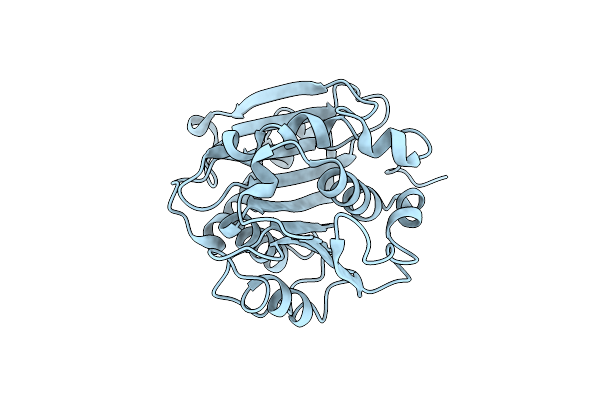 |
Crystal Structure Of A Novel Alpha/Beta Hydrolase From Thermomonospora Curvata In Apo Form
Organism: Thermomonospora curvata dsm 43183
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2023-12-27 Classification: HYDROLASE |
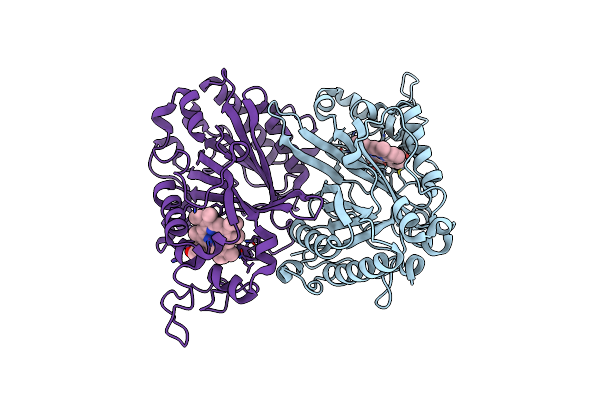 |
Structure Of Thermomonospora Curvata Heme-Containing Dyp-Type Peroxidase E293G Mutant
Organism: Thermomonospora curvata
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2023-11-29 Classification: OXIDOREDUCTASE Ligands: HEM |
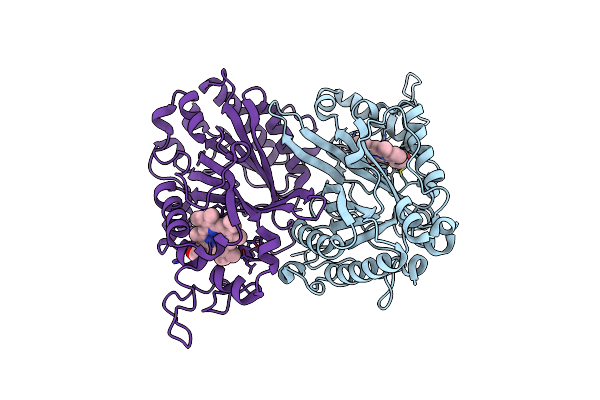 |
Structure Of Thermomonospora Curvata Heme-Containing Dyp-Type Peroxidase E293H Mutant
Organism: Thermomonospora curvata
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2023-11-29 Classification: OXIDOREDUCTASE Ligands: HEM |
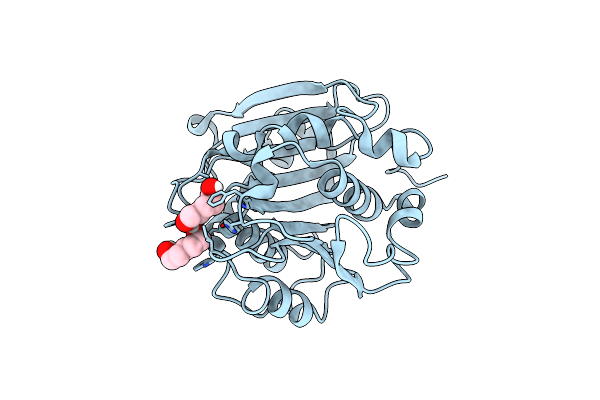 |
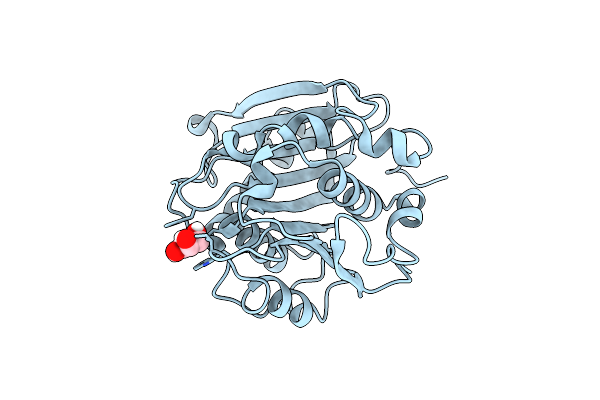
Crystal Structure Of A Novel Alpha/Beta Hydrolase Mutant From Thermomonospora Curvata In Complex With Pentane-1,5-Diol
Organism: Thermomonospora curvata dsm 43183
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2023-07-26 Classification: HYDROLASE Ligands: 9JE |
 |
Crystal Structure Of A Novel Alpha/Beta Hydrolase From Thermomonospora Curvata With Glycerol
Organism: Thermomonospora curvata dsm 43183
Method: X-RAY DIFFRACTION Resolution:1.18 Å Release Date: 2023-07-26 Classification: HYDROLASE Ligands: GOL |
 |
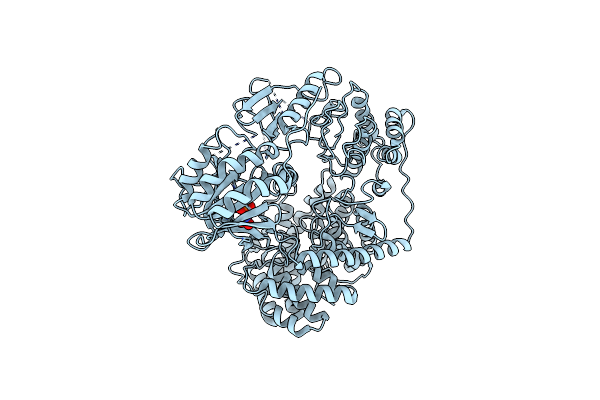
Crystal Structure Of A Novel Alpha/Beta Hydrolase Mutant From Thermomonospora Curvata In Apo Form
Organism: Thermomonospora curvata dsm 43183
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2023-07-26 Classification: HYDROLASE |
 |
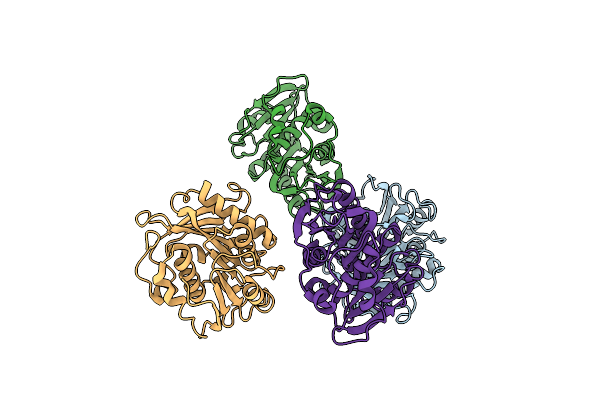
Discovery Of The Lanthipeptide Curvocidin And Structural Insights Into Its Trifunctional Synthetase Cuvl
Organism: Thermomonospora curvata
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2023-06-14 Classification: BIOSYNTHETIC PROTEIN Ligands: PO4, NO3 |
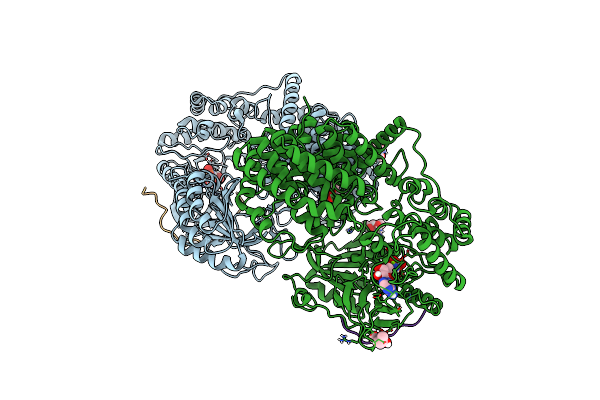 |
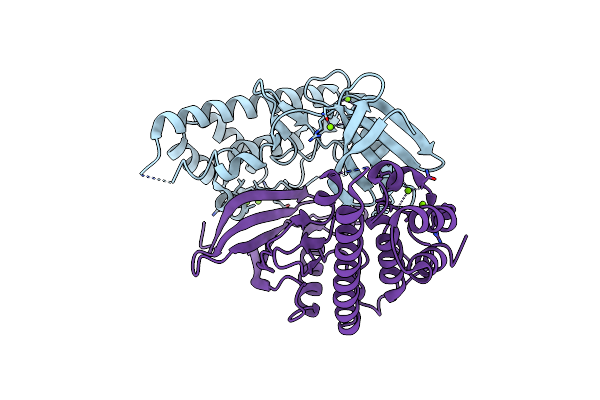
Discovery Of The Lanthipeptide Curvocidin And Structural Insights Into Its Trifunctional Synthetase Cuvl
Organism: Thermomonospora curvata
Method: X-RAY DIFFRACTION Resolution:2.87 Å Release Date: 2023-06-14 Classification: BIOSYNTHETIC PROTEIN Ligands: MPD, ANP, MG |
 |
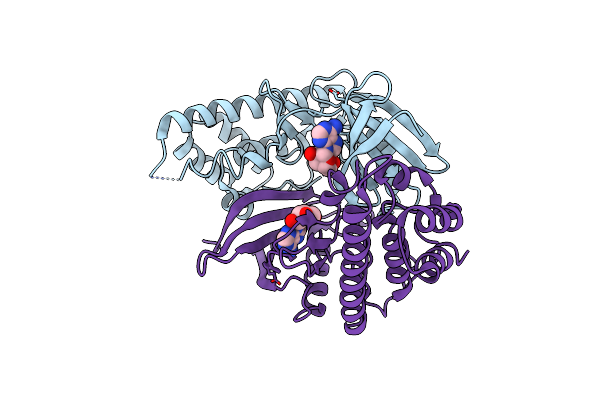
Crystal Structure Of The Kinase Domain Of A Class Iii Lanthipeptide Synthetase Curkc
Organism: Thermomonospora curvata (strain atcc 19995 / dsm 43183 / jcm 3096 / kctc 9072 / nbrc 15933 / ncimb 10081 / henssen b9)
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2022-11-09 Classification: BIOSYNTHETIC PROTEIN Ligands: MG |
 |
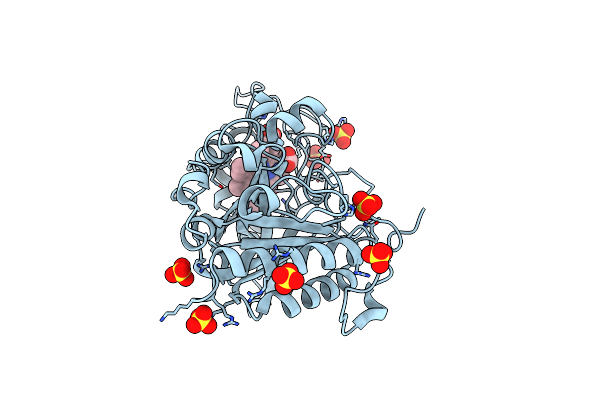
Crystal Structure Of The Kinase Domain With Adenosine Of A Class Iii Lanthipeptide Synthetase Curkc
Organism: Thermomonospora curvata (strain atcc 19995 / dsm 43183 / jcm 3096 / kctc 9072 / nbrc 15933 / ncimb 10081 / henssen b9)
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2022-11-09 Classification: BIOSYNTHETIC PROTEIN Ligands: XYA |
 |
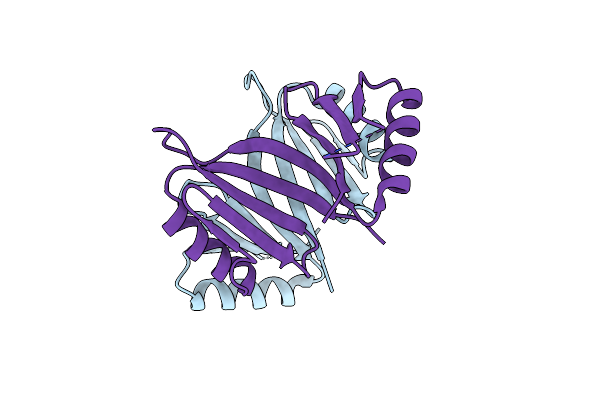
Structure Of Thermomonospora Curvata Heme-Containing Dyp-Type Peroxidase With A Modified Axial Ligand
Organism: Thermomonospora curvata
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-08-31 Classification: OXIDOREDUCTASE Ligands: HEM, SO4 |
 |
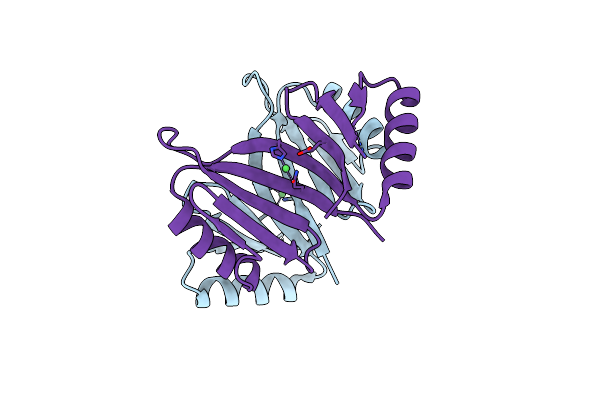
Organism: Thermomonospora curvata (strain atcc 19995 / dsm 43183 / jcm 3096 / nbrc 15933 / ncimb 10081 / henssen b9)
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2021-06-16 Classification: OXIDOREDUCTASE |
 |
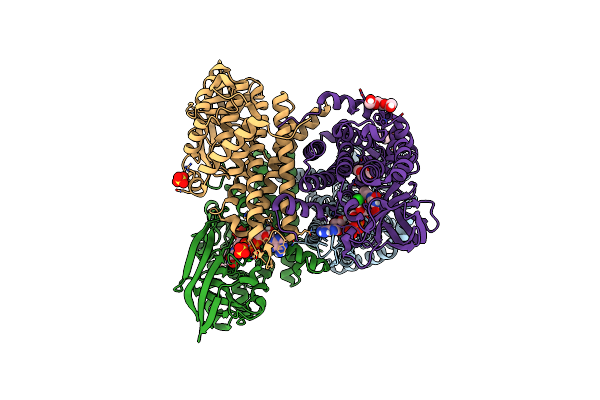
Organism: Thermomonospora curvata (strain atcc 19995 / dsm 43183 / jcm 3096 / nbrc 15933 / ncimb 10081 / henssen b9)
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2021-06-09 Classification: OXIDOREDUCTASE Ligands: NI |
 |
Organism: Thermomonospora curvata (strain atcc 19995 / dsm 43183 / jcm 3096 / nbrc 15933 / ncimb 10081 / henssen b9)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-03-24 Classification: OXIDOREDUCTASE Ligands: PGE, FAD, CL, MES, SO4 |
 |
Crystal Structure Reveals Non-Coordinative Binding Of O2 To The Copper Center Of The Formylglycine-Generating Enzyme - Fge:Ag:S Complex
Organism: Thermomonospora curvata (strain atcc 19995 / dsm 43183 / jcm 3096 / nbrc 15933 / ncimb 10081 / henssen b9), Thermomonospora curvata dsm 43183
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-01-27 Classification: TRANSFERASE Ligands: AG, CA, EDO, CL, SOA |
 |
Crystal Structure Reveals Non-Coordinative Binding Of O2 To The Copper Center Of The Formylglycine-Generating Enzyme - Fge:Ag:S:O2 Complex
Organism: Thermomonospora curvata (strain atcc 19995 / dsm 43183 / jcm 3096 / nbrc 15933 / ncimb 10081 / henssen b9), Thermomonospora curvata dsm 43183
Method: X-RAY DIFFRACTION Release Date: 2021-01-27 Classification: TRANSFERASE Ligands: OXY, CA, CL, AG, SOA |
 |
Crystal Structure Reveals Non-Coordinative Binding Of O2 To The Copper Center Of The Formylglycine-Generating Enzyme - Fge:Ag:S:No Complex
Organism: Thermomonospora curvata (strain atcc 19995 / dsm 43183 / jcm 3096 / nbrc 15933 / ncimb 10081 / henssen b9), Thermomonospora curvata dsm 43183
Method: X-RAY DIFFRACTION Release Date: 2021-01-27 Classification: TRANSFERASE Ligands: AG, CA, CL, NO, EDO, SOA |
 |
Crystal Structure Reveals Non-Coordinative Binding Of O2 To The Copper Center Of The Formylglycine-Generating Enzyme - Fge:Cu:S:No Complex
Organism: Thermomonospora curvata (strain atcc 19995 / dsm 43183 / jcm 3096 / nbrc 15933 / ncimb 10081 / henssen b9), Thermomonospora curvata dsm 43183
Method: X-RAY DIFFRACTION Release Date: 2021-01-27 Classification: TRANSFERASE Ligands: CU1, CA, NO, SOA |
 |
Crystal Structure Reveals Non-Coordinative Binding Of O2 To The Copper Center Of The Formylglycine-Generating Enzyme - Fge:Cu:S:O2-1A Complex
Organism: Thermomonospora curvata (strain atcc 19995 / dsm 43183 / jcm 3096 / nbrc 15933 / ncimb 10081 / henssen b9), Thermomonospora curvata dsm 43183
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-01-27 Classification: TRANSFERASE Ligands: CU1, CA, OXY, SOA |
 |
Crystal Structure Reveals Non-Coordinative Binding Of O2 To The Copper Center Of The Formylglycine-Generating Enzyme - Fge:Cu:S:O2-1B Complex
Organism: Thermomonospora curvata (strain atcc 19995 / dsm 43183 / jcm 3096 / nbrc 15933 / ncimb 10081 / henssen b9), Thermomonospora curvata dsm 43183
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2021-01-27 Classification: TRANSFERASE Ligands: CU1, CA, OXY, CL, MG, SOA |

