Search Count: 34
 |
Organism: Escherichia coli (strain k12), Rhodospirillum rubrum (strain atcc 11170 / ath 1.1.1 / dsm 467 / lmg 4362 / ncimb 8255 / s1)
Method: ELECTRON MICROSCOPY Release Date: 2025-02-12 Classification: CHAPERONE Ligands: AF3, MG, ADP, K |
 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2023-12-27 Classification: CHAPERONE Ligands: BEF, ADP, MG, K |
 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2023-12-27 Classification: CHAPERONE Ligands: AF3, ADP, MG, K |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-10-25 Classification: CHAPERONE |
 |
Organism: Methanocaldococcus jannaschii
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2023-07-12 Classification: DNA BINDING PROTEIN |
 |
Organism: Human herpesvirus 8
Method: X-RAY DIFFRACTION Resolution:4.20 Å Release Date: 2022-05-11 Classification: VIRAL PROTEIN |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2019-01-23 Classification: TRANSFERASE |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2019-01-23 Classification: TRANSFERASE Ligands: APC, MN |
 |
Crystal Structure Of Daca From Staphylococcus Aureus, N166C/T172C Double Mutant
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.77 Å Release Date: 2019-01-23 Classification: TRANSFERASE |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2019-01-23 Classification: ISOMERASE |
 |
Organism: Thrixopelma pruriens
Method: X-RAY DIFFRACTION Resolution:0.99 Å Release Date: 2017-09-13 Classification: TOXIN Ligands: CL, EDO |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.82 Å Release Date: 2017-02-08 Classification: TRANSCRIPTION |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:1.21 Å Release Date: 2015-07-08 Classification: LYASE Ligands: NAG, DMS, 3P9 |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2015-07-08 Classification: LYASE Ligands: EDO |
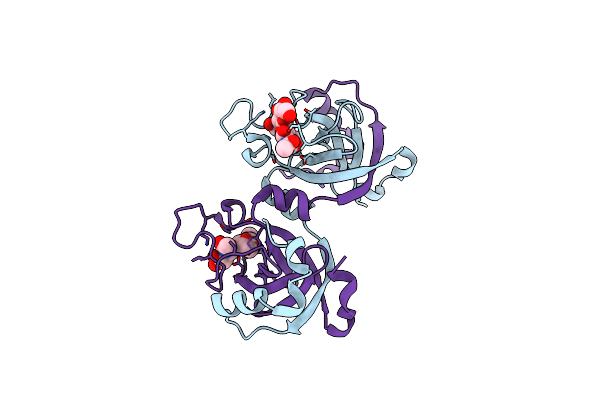 |
Stationary Phase Survival Protein Yuic From B.Subtilis Complexed With Reaction Product
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2015-07-08 Classification: LYASE Ligands: 3QL |
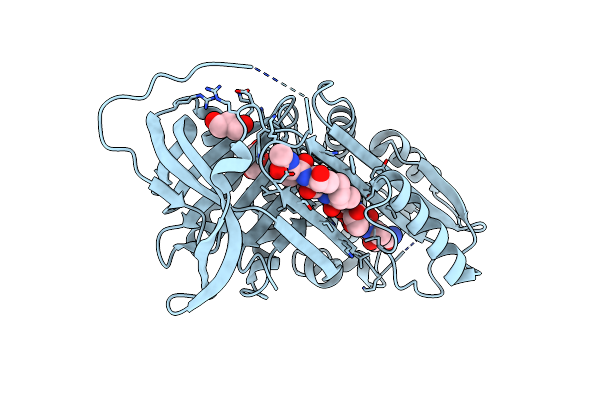 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2015-06-10 Classification: Hydrolase inhibitor Ligands: GOL |
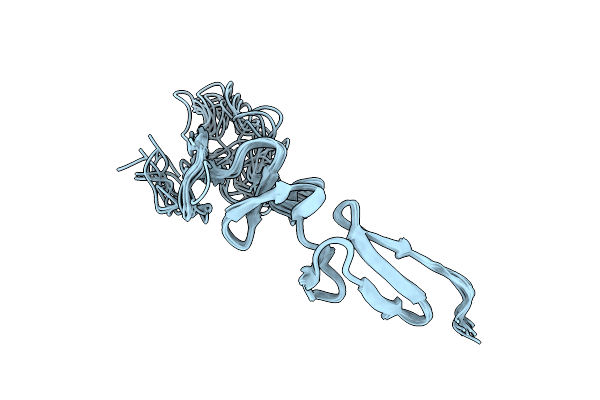 |
Solution Structure Of The Major Factor Viii Binding Region On Von Willebrand Factor
|
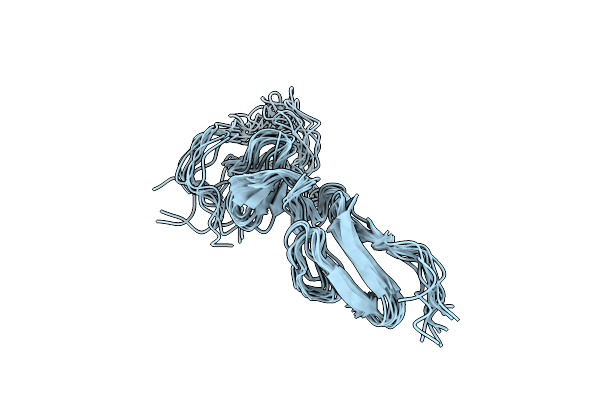 |
Solution Structure Of The Major Factor Viii Binding Region On Von Willebrand Factor
|
 |
Organism: Actinidia deliciosa
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2013-09-04 Classification: ALLERGEN Ligands: UNL, CL |
 |
Organism: Actinidia deliciosa
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2013-09-04 Classification: ALLERGEN Ligands: UNL, CL, EDO |

