Search Count: 17
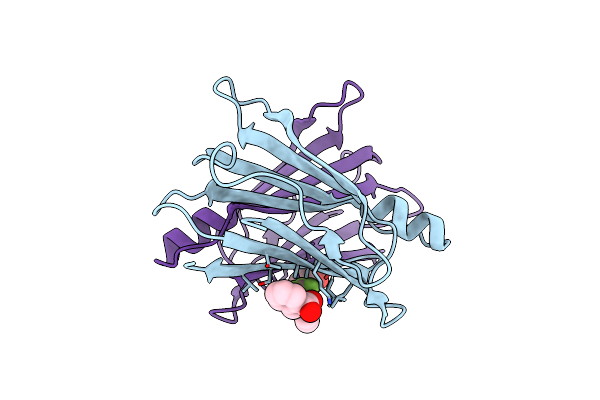 |
Human Transthyretin (Ttr) In Complex With (E)-2-((((2-(Trifluoromethyl)Benzyl)Oxy)Imino)Methyl)Benzoic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: PROTEIN TRANSPORT Ligands: A1IC9 |
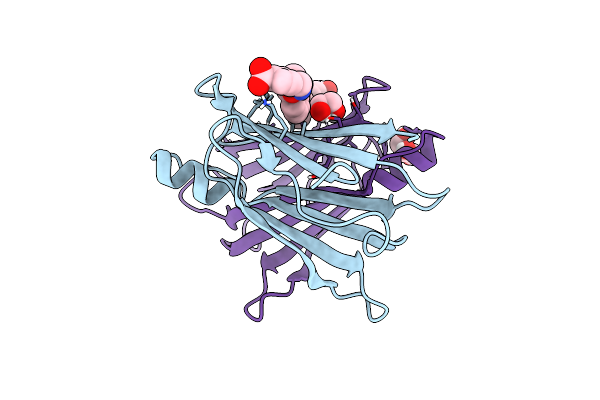 |
Human Transthyretin (Ttr) In Complex With (E)-4-((((2-Methoxybenzyl)Oxy)Imino)Methyl)Benzoic Acid (Lic157)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-04 Classification: PROTEIN TRANSPORT Ligands: A1ICE, GOL, EDO |
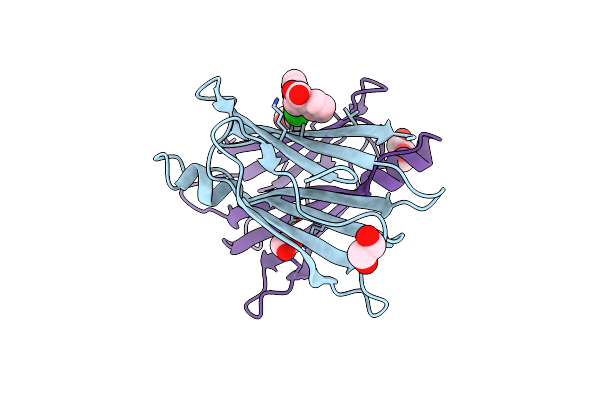 |
Human Transthyretin (Ttr) In Complex With (E)-2-((((2-Chlorobenzyl)Oxy)Imino)Methyl)Benzoic Acid (Lic166)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-04 Classification: PROTEIN TRANSPORT Ligands: EDO, A1ICF, GOL |
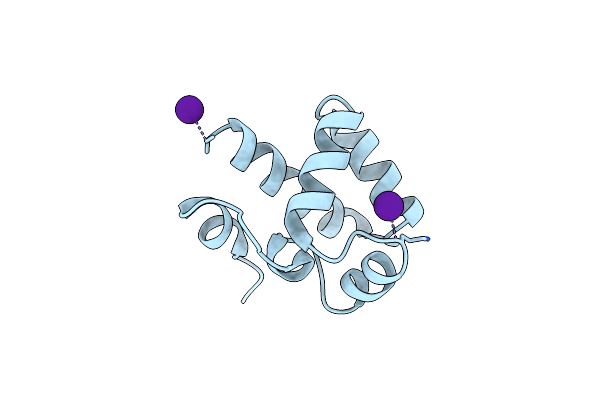 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2021-07-07 Classification: DNA BINDING PROTEIN Ligands: CS |
 |
Di-Phosphorylated Barrier-To-Autointegration Factor (Baf) In Complex With Lem Domain Of Emerin
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2021-04-07 Classification: DNA BINDING PROTEIN Ligands: EPE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2016-02-10 Classification: CELL CYCLE Ligands: GOL, MG |
 |
Organism: Streptomyces noursei
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2014-10-08 Classification: transferase/transferase inhibitor Ligands: XVE |
 |
Substrate And Reaction Specificity Of Mycobacterium Tuberculosis Cytochrome P450 Cyp121
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2013-05-01 Classification: OXIDOREDUCTASE Ligands: SO4, HEM, 1ED |
 |
Substrate And Reaction Specificity Of Mycobacterium Tuberculosis Cytochrome P450 Cyp121
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2013-05-01 Classification: OXIDOREDUCTASE Ligands: HEM, SO4, 1G4, GOL |
 |
Substrate And Reaction Specificity Of Mycobacterium Tuberculosis Cytochrome P450 Cyp121
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2013-05-01 Classification: OXIDOREDUCTASE Ligands: HEM, SO4, 1G7, GOL, PO4 |
 |
Substrate And Reaction Specificity Of Mycobacterium Tuberculosis Cytochrome P450 Cyp121
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-05-01 Classification: OXIDOREDUCTASE Ligands: HEM, SO4, 1G9 |
 |
Substrate And Reaction Specificity Of Mycobacterium Tuberculosis Cytochrome P450 Cyp121
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2013-05-01 Classification: OXIDOREDUCTASE Ligands: HEM, SO4, GOL, 1GB |
 |
Organism: Streptomyces noursei
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2011-02-16 Classification: PROTEIN BINDING Ligands: DTD, PO4 |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2010-12-15 Classification: ISOMERASE/INHIBITOR Ligands: RAP |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2009-04-21 Classification: OXIDOREDUCTASE Ligands: SO4, HEM |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2009-04-21 Classification: OXIDOREDUCTASE Ligands: HEM, YTT, SO4 |
 |
Organism: Corynespora cassiicola
Method: SOLUTION NMR Release Date: 2007-02-27 Classification: TOXIN Ligands: 3HD |

