Search Count: 184
 |
Organism: Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: METAL BINDING PROTEIN Ligands: CO, CA, CL, NA |
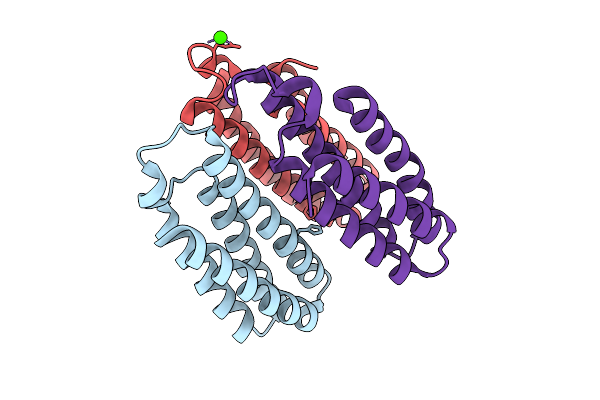 |
Organism: Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: METAL BINDING PROTEIN Ligands: NI, CL, CA |
 |
Organism: Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: METAL BINDING PROTEIN |
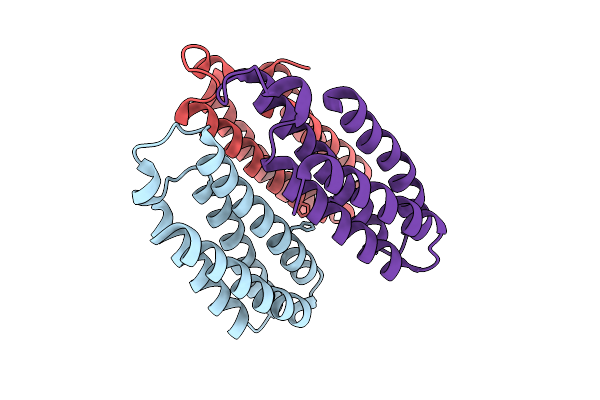 |
Organism: Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: METAL BINDING PROTEIN Ligands: CU |
 |
Organism: Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: METAL BINDING PROTEIN Ligands: ZN, CA, NA |
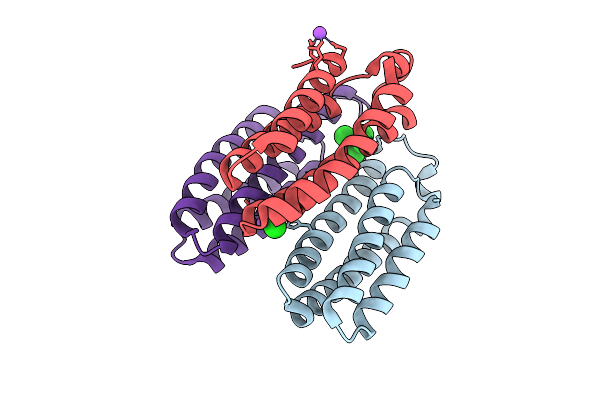 |
Organism: Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: METAL BINDING PROTEIN Ligands: ZN, CL, BS3, NA |
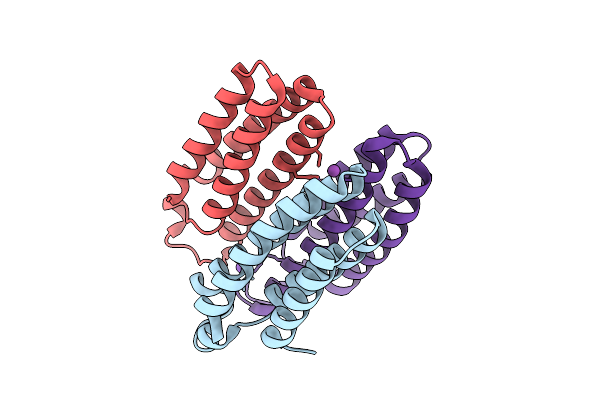 |
Organism: Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: METAL BINDING PROTEIN Ligands: BS3, NA, CO |
 |
Organism: Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: METAL BINDING PROTEIN Ligands: BS3, NI, CA |
 |
Organism: Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: METAL BINDING PROTEIN Ligands: CL, NA, BS3 |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: DE NOVO PROTEIN Ligands: ER3, CL |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: DE NOVO PROTEIN Ligands: CU |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: DE NOVO PROTEIN Ligands: RH |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: DE NOVO PROTEIN Ligands: CU, CA |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: DE NOVO PROTEIN Ligands: NI |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: DE NOVO PROTEIN Ligands: CO |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: DE NOVO PROTEIN Ligands: FE, CA |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: DE NOVO PROTEIN Ligands: MN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: DE NOVO PROTEIN Ligands: ZN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: DE NOVO PROTEIN Ligands: ZN |

