Search Count: 17
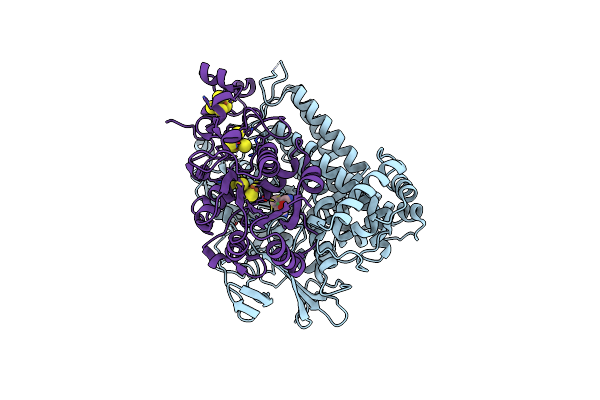 |
Crystal Structure Of The C120G Variant Of The Membrane-Bound [Nife]-Hydrogenase From Cupriavidus Necator In The Air-Oxidized State At 1.93 A Resolution.
Organism: Cupriavidus necator h16
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2025-02-26 Classification: OXIDOREDUCTASE Ligands: NFV, MG, SF4, F3S, 35L, SF3, CL |
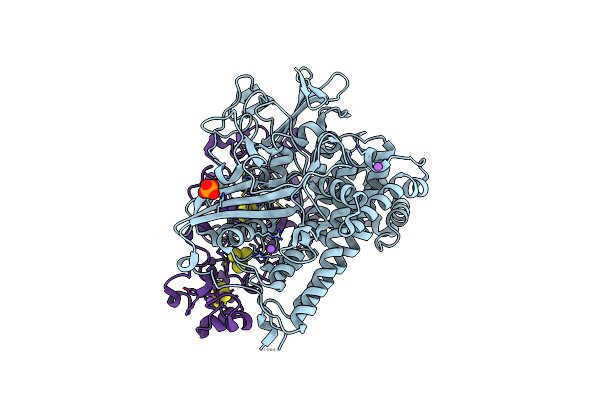 |
Crystal Structure Of The C19G Variant Of The Membrane-Bound [Nife]-Hydrogenase From Cupriavidus Necator In The H2-Reduced State At 1.6 A Resolution.
Organism: Cupriavidus necator h16
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-09-18 Classification: OXIDOREDUCTASE Ligands: NFU, MG, NA, PO4, SF4, F3S, CL, ER2 |
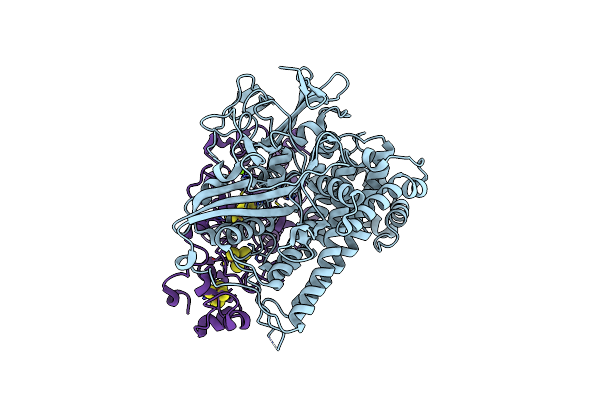 |
Crystal Structure Of The C120G Variant Of The Membrane-Bound [Nife]-Hydrogenase From Cupriavidus Necator In The H2-Reduced State At 1.65 A Resolution.
Organism: Cupriavidus necator h16
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2024-09-18 Classification: OXIDOREDUCTASE Ligands: NFU, MG, CL, SF4, F3S, 35L, F4S |
 |
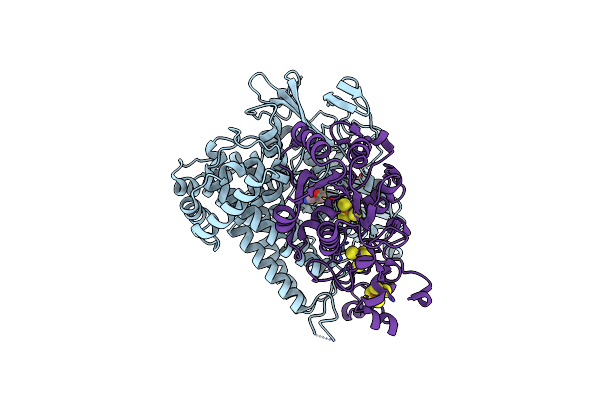
Crystal Structure Of The C19G Variant Of The Membrane-Bound [Nife]-Hydrogenase From Cupriavidus Necator In The Air-Oxidized State At 1.61 A Resolution.
Organism: Cupriavidus necator h16
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2023-11-15 Classification: OXIDOREDUCTASE Ligands: NFV, MG, CL, SF4, F3S, ER2 |
 |
Crystal Structure Of The C19G/C120G Variant Of The Membrane-Bound [Nife]-Hydrogenase From Cupriavidus Necator In The Air-Oxidized State At 1.65 A Resolution.
Organism: Cupriavidus necator h16
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-11-01 Classification: OXIDOREDUCTASE Ligands: NFV, MG, CL, SF4, F3S |
 |
Crystal Structure Of The C19G/C120G Variant Of The Membrane-Bound [Nife]-Hydrogenase From Cupriavidus Necator In The H2-Reduced State At 1.92 A Resolution.
Organism: Cupriavidus necator h16
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2023-11-01 Classification: OXIDOREDUCTASE Ligands: NFU, MG, SF4, F3S |
 |
Organism: Carboxydothermus hydrogenoformans (strain atcc baa-161 / dsm 6008 / z-2901)
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2022-01-12 Classification: OXIDOREDUCTASE Ligands: FES, SF4, T2N, TAM, PEG |
 |
Organism: Carboxydothermus hydrogenoformans (strain atcc baa-161 / dsm 6008 / z-2901)
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2022-01-12 Classification: OXIDOREDUCTASE Ligands: FES, SF4, SF3, BU3, PEG |
 |
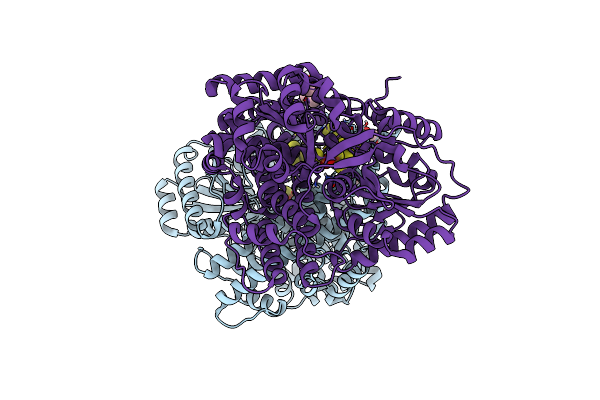
Organism: Carboxydothermus hydrogenoformans (strain atcc baa-161 / dsm 6008 / z-2901)
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2022-01-12 Classification: OXIDOREDUCTASE Ligands: FES, SF4, SF3, BU3, PEG, TAM |
 |
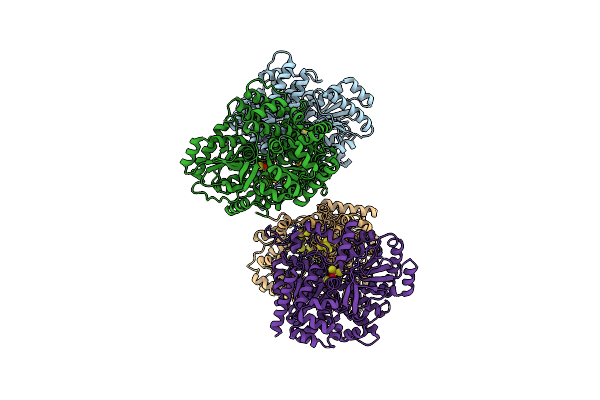
Organism: Carboxydothermus hydrogenoformans (strain atcc baa-161 / dsm 6008 / z-2901)
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2022-01-12 Classification: OXIDOREDUCTASE Ligands: FES, SF4, T2N, H2S, BU3 |
 |
Organism: Carboxydothermus hydrogenoformans (strain atcc baa-161 / dsm 6008 / z-2901)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2022-01-12 Classification: OXIDOREDUCTASE Ligands: SF4, XE, BR, T2N, H2S, FES |
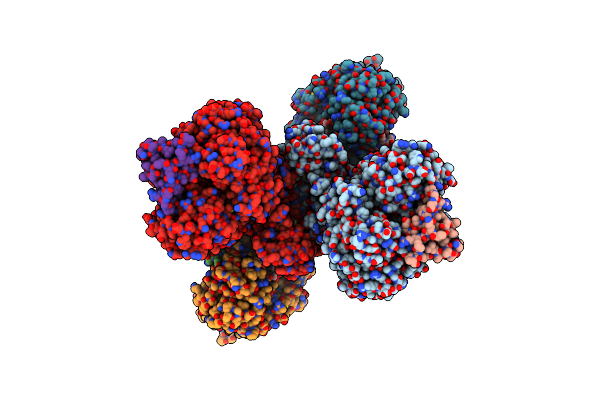 |
Cryo-Em Structure Of Nadh Reduced Form Of Nad+-Dependent Formate Dehydrogenase From Rhodobacter Capsulatus
Organism: Rhodobacter capsulatus
Method: ELECTRON MICROSCOPY Resolution:3.24 Å Release Date: 2020-04-22 Classification: OXIDOREDUCTASE Ligands: MGD, 6MO, FES, SF4, H2S, FMN, NAI |
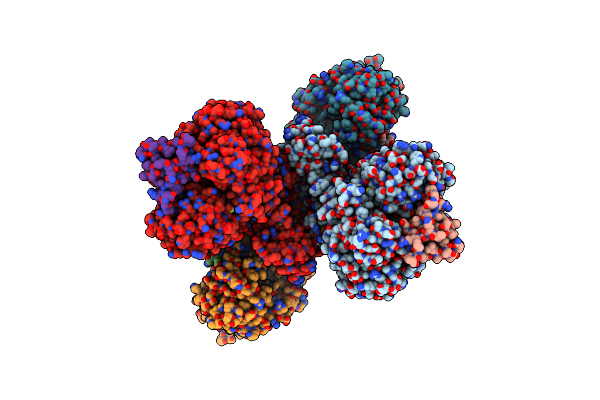 |
Cryo-Em Structure Of As Isolated Form Of Nad+-Dependent Formate Dehydrogenase From Rhodobacter Capsulatus
Organism: Rhodobacter capsulatus
Method: ELECTRON MICROSCOPY Resolution:3.26 Å Release Date: 2020-04-22 Classification: OXIDOREDUCTASE Ligands: MGD, 6MO, FES, SF4, H2S, FMN |
 |
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.18 Å Release Date: 2017-03-08 Classification: HYDROLASE Ligands: RXR, CL, HED, PO4, BME, K |
 |
Crystal Structure Of An O2-Tolerant [Nife]-Hydrogenase From Ralstonia Eutropha In Its As-Isolated Form - Oxidized State 1
Organism: Ralstonia eutropha
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2014-04-02 Classification: OXIDOREDUCTASE Ligands: NFV, MG, PO4, SF4, F3S, S3F, CL |
 |
Crystal Structure Of An O2-Tolerant [Nife]-Hydrogenase From Ralstonia Eutropha In Its As-Isolated Form - Oxidized State 2
Organism: Ralstonia eutropha
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2014-04-02 Classification: OXIDOREDUCTASE Ligands: NFV, MG, CL, SF4, F3S, S3F |
 |
Crystal Structure Of An O2-Tolerant [Nife]-Hydrogenase From Ralstonia Eutropha In Its As-Isolated Form With Ascorbate - Partly Reduced State
Organism: Ralstonia eutropha
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2014-04-02 Classification: OXIDOREDUCTASE Ligands: NFV, MG, SF4, F3S, S3F, CL |

