Search Count: 31
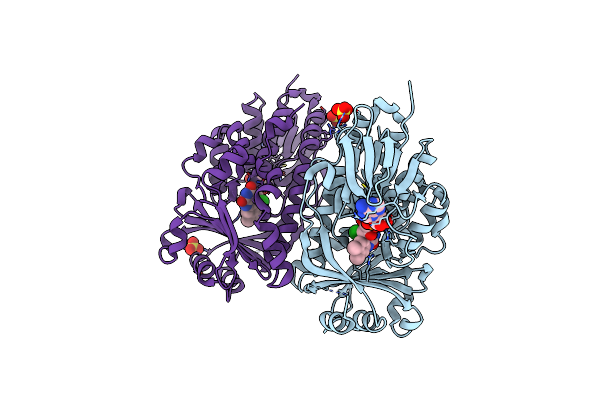 |
Crystal Structure Of The Flavoprotein Monooxygenase Trle From Streptomyces Cyaneofuscatus Soc7
Organism: Streptomyces cyaneofuscatus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-05-15 Classification: FLAVOPROTEIN Ligands: FAD, SO4, CL |
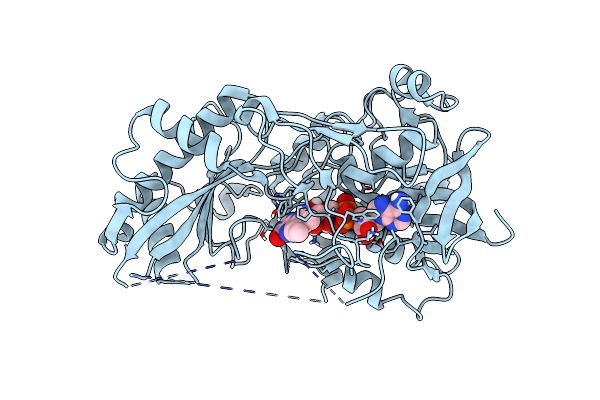 |
Crystal Structure Of The Chloramphenicol-Inactivating Oxidoreductase From Novosphingobium Sp
Organism: Novosphingobium sp. b 225
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-11-16 Classification: FLAVOPROTEIN Ligands: FAD |
 |
Crystal Structure Of The Flavoprotein Monooxygenase Grho5 From Griseorhodin A Biosynthesis
Organism: Streptomyces sp. jp95
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2021-11-03 Classification: FLAVOPROTEIN Ligands: FAD |
 |
Crystal Structure Of A Ternary Complex Of The Flavoprotein Monooxygenase Grho5 With Fad And Collinone
Organism: Streptomyces sp. jp95
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2021-11-03 Classification: FLAVOPROTEIN Ligands: FAD, 1J0 |
 |
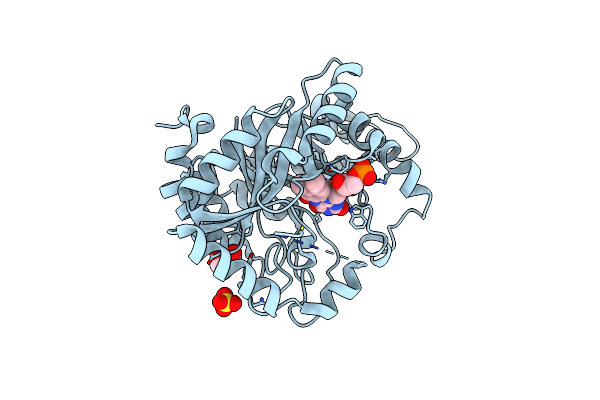
Crystal Structure Of The Flavoprotein Monooxygenase Rubl From Rubromycin Biosynthesis
Organism: Streptomyces collinus
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2021-11-03 Classification: FLAVOPROTEIN Ligands: FAD, CL, 1JW, HYP, GOL |
 |
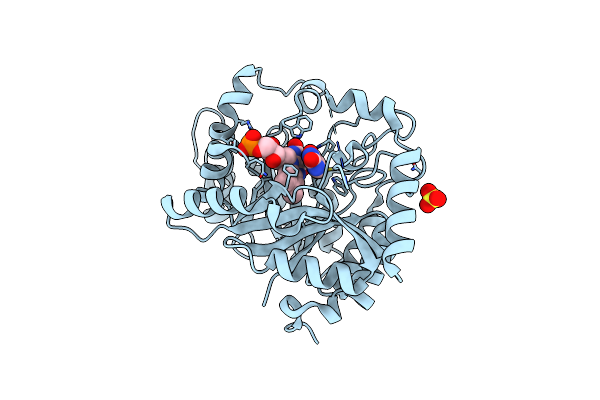
Crystal Structure Of Monooxygenase Ruta Complexed With Dioxygen Under 1.5 Mpa / 15 Bars Of Oxygen Pressure.
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-02-05 Classification: FLAVOPROTEIN Ligands: SO4, GOL, FMN, OXY |
 |
Crystal Structure Of Monooxygenase Ruta Complexed With Uracil Under Atmospheric Pressure.
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2020-02-05 Classification: FLAVOPROTEIN Ligands: FMN, URA, SO4 |
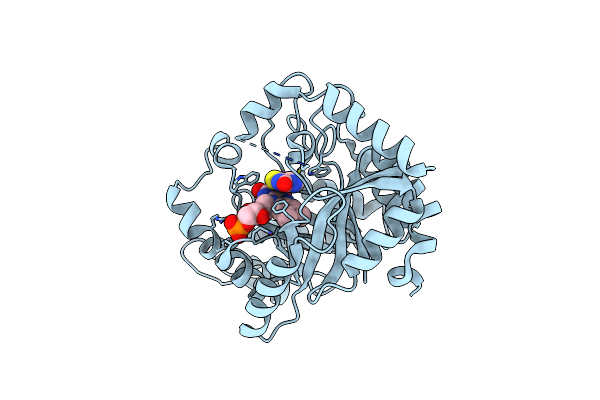 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-02-05 Classification: FLAVOPROTEIN Ligands: FMN, LDB |
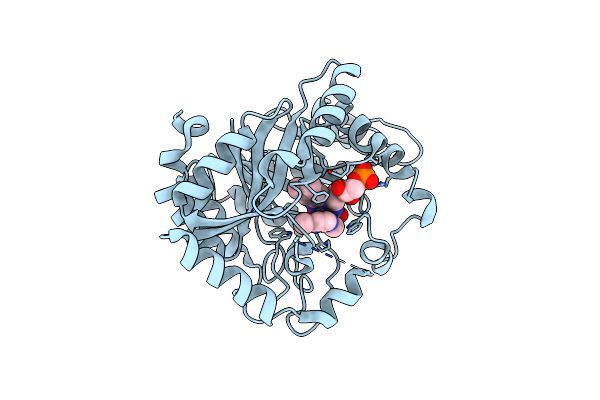 |
Crystal Structure Of Monooxygenase Ruta Complexed With 2,4-Dimethoxypyrimidine.
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2020-02-05 Classification: FLAVOPROTEIN Ligands: FMN, LD8 |
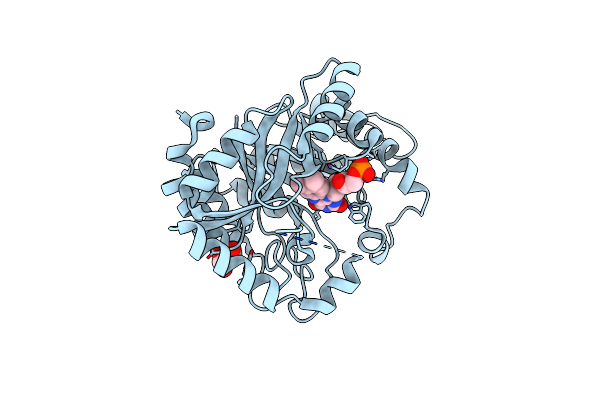 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-02-05 Classification: FLAVOPROTEIN Ligands: GOL, FMN |
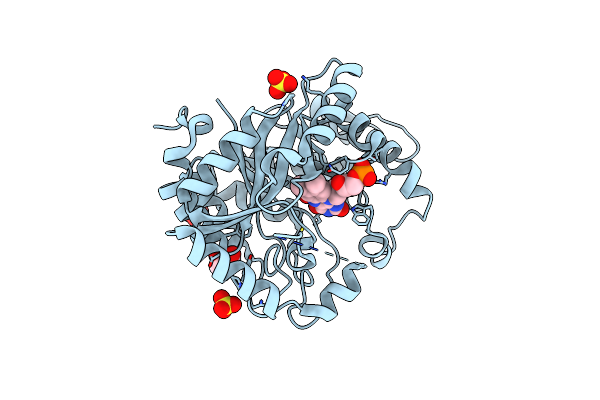 |
Crystal Structure Of Monooxygenase Ruta Complexed With Dioxygen Under 0.5 Mpa / 5 Bars Of Oxygen Pressure.
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-02-05 Classification: FLAVOPROTEIN Ligands: SO4, GOL, FMN, OXY |
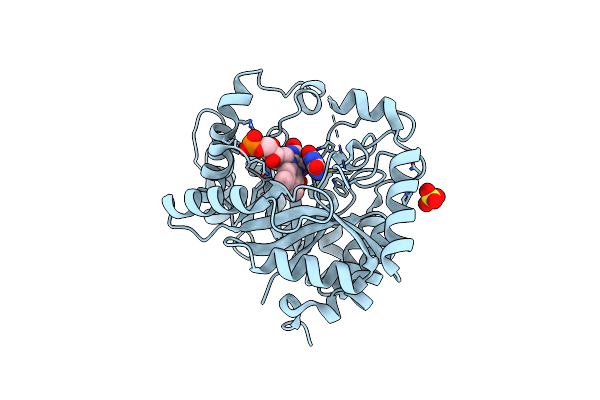 |
Crystal Structure Of Monooxygenase Ruta Complexed With Uracil And Dioxygen Under 1.5 Mpa / 15 Bars Of Oxygen Pressure.
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-02-05 Classification: FLAVOPROTEIN Ligands: FMN, URA, SO4, OXY |
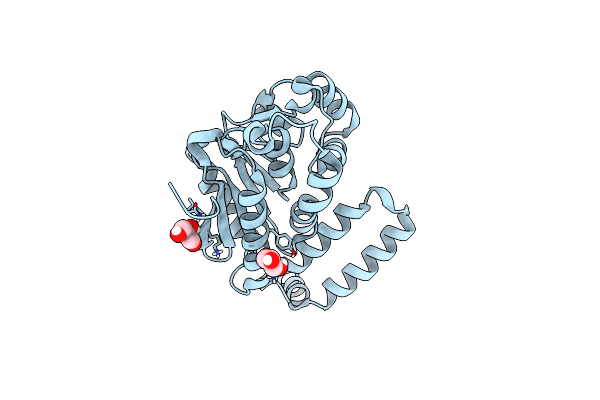 |
Organism: Thermus thermophilus jl-18
Method: X-RAY DIFFRACTION Resolution:1.27 Å Release Date: 2019-12-11 Classification: ISOMERASE Ligands: GOL |
 |
Organism: Thermus thermophilus jl-18
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2019-12-11 Classification: ISOMERASE Ligands: LHQ |
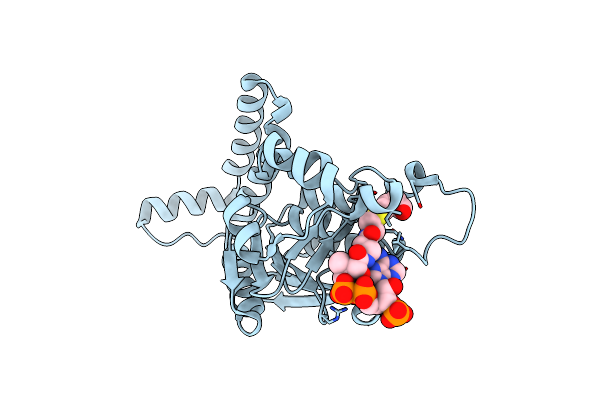 |
Organism: Thermus thermophilus jl-18
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2019-12-11 Classification: ISOMERASE Ligands: T3D |
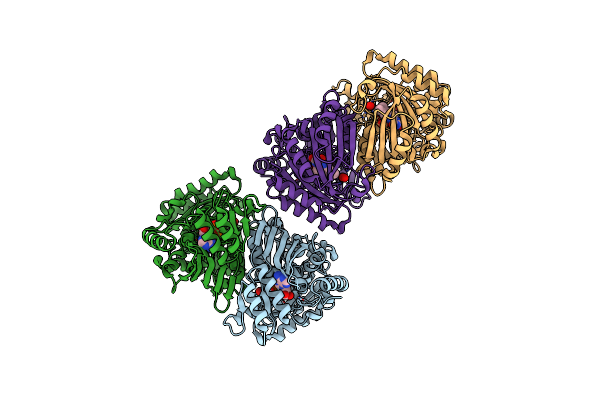 |
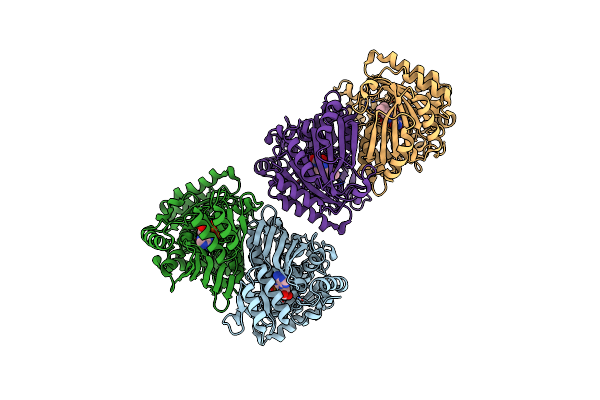
The Crystal Structure Of Encm Complexed With Dioxygen Under 15 Bar Of Oxygen Pressure.
Organism: Streptomyces maritimus
Method: X-RAY DIFFRACTION Resolution:1.39 Å Release Date: 2018-05-02 Classification: FLAVOPROTEIN Ligands: FAD, OXY |
 |
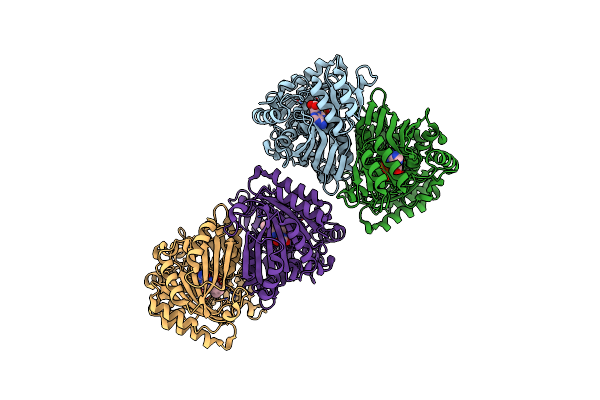
The Crystal Structure Of Encm Complexed With Dioxygen Under 10 Bar Of Oxygen Pressure.
Organism: Streptomyces maritimus
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2018-05-02 Classification: FLAVOPROTEIN Ligands: FAD, OXY |
 |
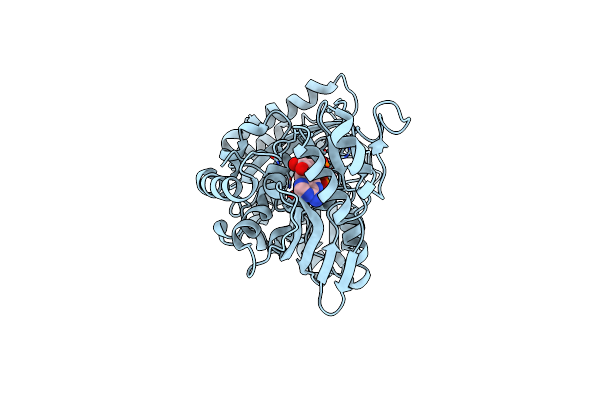
The Crystal Structure Of Encm Complexed With Dioxygen Under 5 Bar Of Oxygen Pressure.
Organism: Streptomyces maritimus
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2018-05-02 Classification: FLAVOPROTEIN Ligands: FAD, OXY |
 |
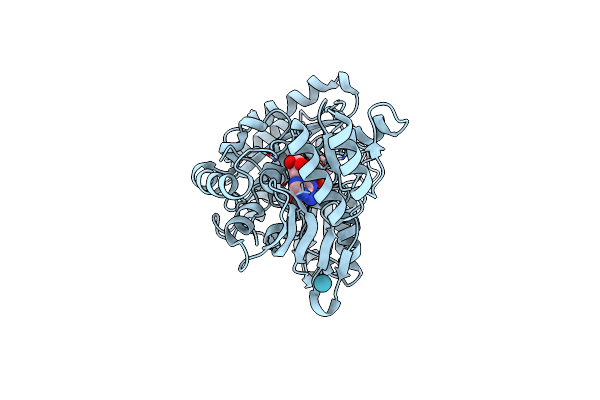
Organism: Streptomyces maritimus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2018-05-02 Classification: FLAVOPROTEIN Ligands: FAD, BR |
 |
Organism: Streptomyces maritimus
Method: X-RAY DIFFRACTION Resolution:3.65 Å Release Date: 2018-05-02 Classification: FLAVOPROTEIN Ligands: FAD, XE |

