Search Count: 28
 |
Structure Of Sars-3Cl Protease Complex With A Phenyl-Beta-Alanyl (R,S)-N-Decalin Type Inhibitor
Organism: Human sars coronavirus
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2016-06-22 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: SLH |
 |
Structure Of Sars-3Cl Protease Complex With A Phenyl-Beta-Alanyl (S,R)-N-Decalin Type Inhibitor
Organism: Human sars coronavirus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2016-06-22 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: SDJ |
 |
Synthesis, Carbonic Anhydrase Inhibition And Protein X-Ray Structure Of The Unusual Natural Product Primary Sulfonamide Psammaplin C
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2016-05-25 Classification: LYASE Ligands: ZN, DMS, OE2 |
 |
An Unusual Natural Product Primary Sulfonamide: Synthesis, Carbonic Anhydrase Inhibition And Protein X-Ray Structure Of Psammaplin C
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2016-05-25 Classification: LYASE Ligands: ZN, OE2, SO4, NA |
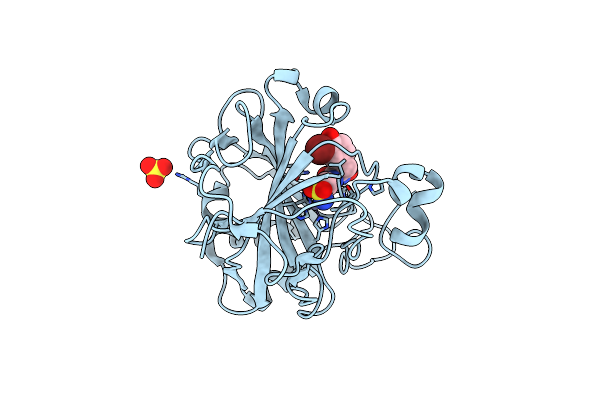 |
An Unusual Natural Product Primary Sulfonamide: Synthesis, Carbonic Anhydrase Inhibition And Protein X-Ray Structure Of Psammaplin C
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2016-05-25 Classification: LYASE Ligands: ZN, SO4, OE2 |
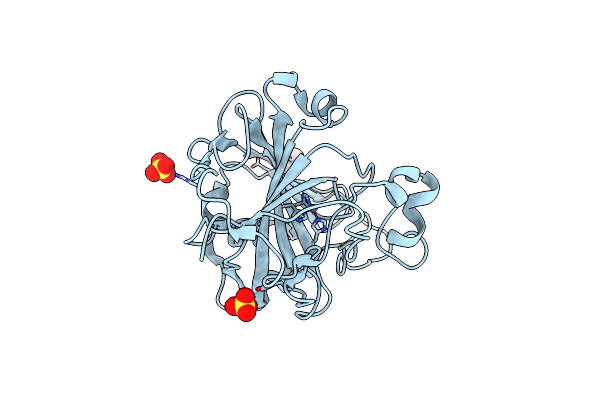 |
An Unusual Natural Product Primary Sulfonamide: Synthesis, Carbonic Anhydrase Inhibition And Protein X-Ray Structure Of Psammaplin C
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2016-05-25 Classification: LYASE Ligands: ZN, SO4, GOL, CL |
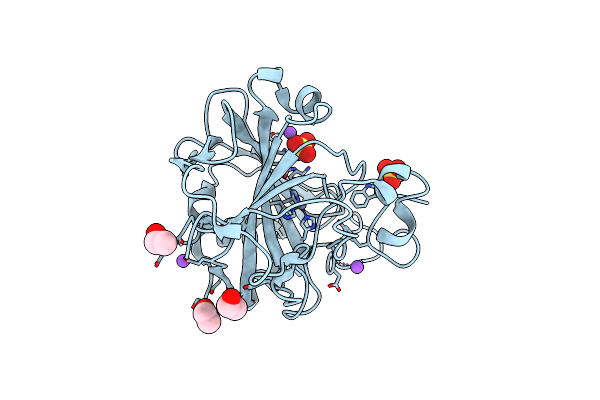 |
An Unusual Natural Product Primary Sulfonamide: Synthesis, Carbonic Anhydrase Inhibition And Protein X-Ray Structure Of Psammaplin C
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.28 Å Release Date: 2016-05-25 Classification: LYASE Ligands: ZN, NA, SO4, EOH |
 |
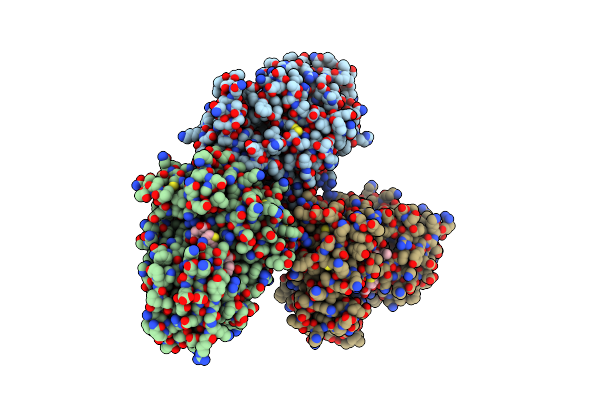
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2015-07-29 Classification: HYDROLASE/HYDROLASE INHIBITOR |
 |
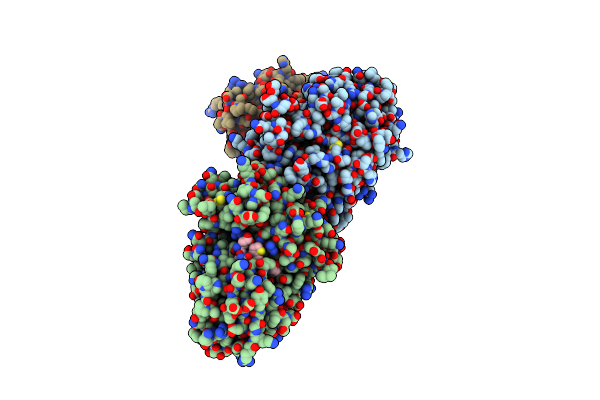
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2015-07-01 Classification: Hydrolase/Hydrolase inhibitor |
 |
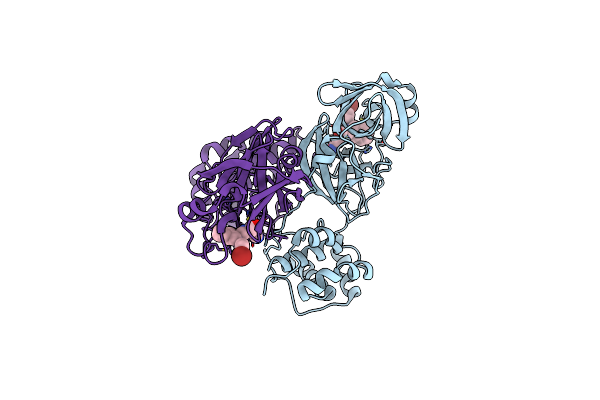
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.25 Å Release Date: 2015-07-01 Classification: Hydrolase/Hydrolase inhibitor |
 |
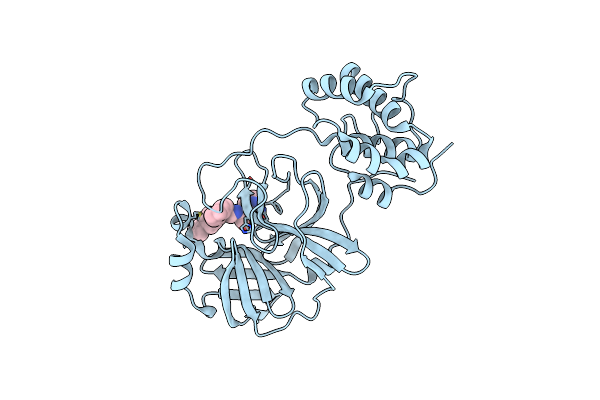
Structure Of Sars-3Cl Protease Complex With A Bromobenzoyl (S,R)-N-Decalin Type Inhibitor
Organism: Human sars coronavirus
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2015-02-18 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 3A7 |
 |
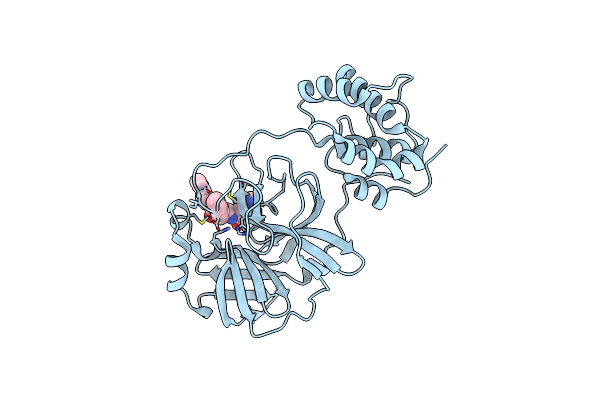
Structure Of Sars-3Cl Protease Complex With A Phenylbenzoyl (S,R)-N-Decalin Type Inhibitor
Organism: Human sars coronavirus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2015-02-18 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 3BL |
 |
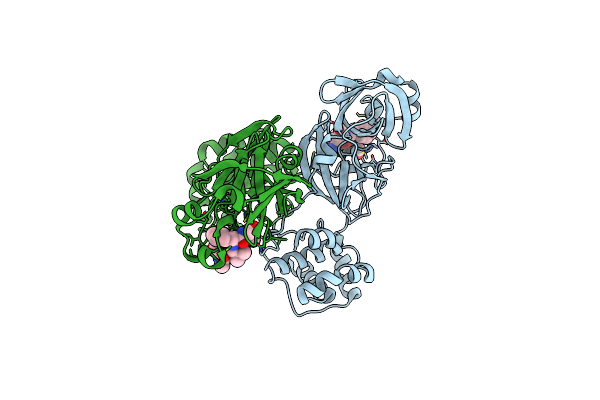
Structure Of Sars-3Cl Protease Complex With A Phenylbenzoyl (R,S)-N-Decalin Type Inhibitor
Organism: Sars coronavirus
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2015-02-18 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 3X5 |
 |
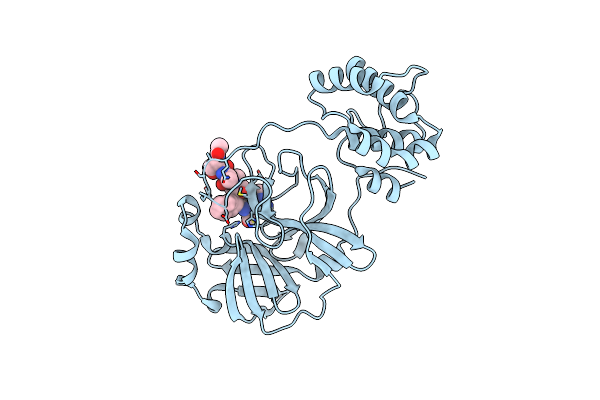
Structure-Based Design, Synthesis, Evaluation Of Peptide-Mimetic Sars 3Cl Protease Inhibitors
Organism: Sars coronavirus
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2011-12-14 Classification: HYDROLASE/HYDROLASE INHIBITOR |
 |
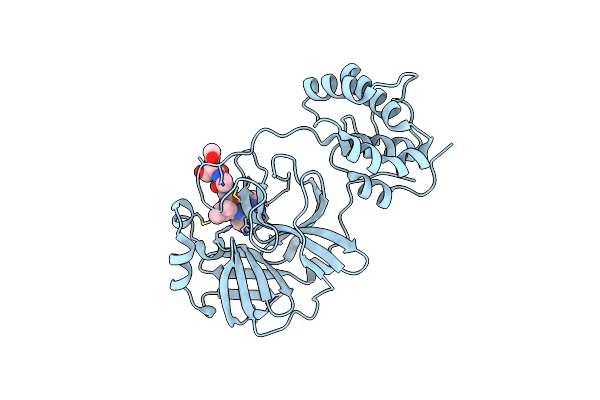
Structure Of Sars 3Cl Protease With Peptidic Aldehyde Inhibitor Containing Cyclohexyl Side Chain
Organism: Sars coronavirus
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2011-12-14 Classification: HYDROLASE/HYDROLASE inhibitor |
 |
Organism: Sars coronavirus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2011-12-14 Classification: HYDROLASE/HYDROLASE inhibitor |
 |
Structure Of Sars 3Cl Protease Auto-Proteolysis Resistant Mutant In The Absent Of Inhibitor
Organism: Sars coronavirus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-12-14 Classification: HYDROLASE |
 |
Crystal Structure Of Human Gluk2 Ligand-Binding Core In Complex With Novel Marine-Derived Toxins, Neodysiherbaine A
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2011-10-26 Classification: MEMBRANE PROTEIN Ligands: NDZ |
 |
Crystal Structure Of The Human Glutamate Receptor, Glur5, Ligand-Binding Core In Complex With L-Glutamate In Space Group P1
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2010-01-19 Classification: MEMBRANE PROTEIN Ligands: GLU, SO4 |
 |
Crystal Structure Of The Human Glutamate Receptor, Glur5, Ligand-Binding Core In Complex With Dysiherbaine In Space Group P1
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2010-01-19 Classification: MEMBRANE PROTEIN Ligands: DYH, SO4, GOL |

