Search Count: 8
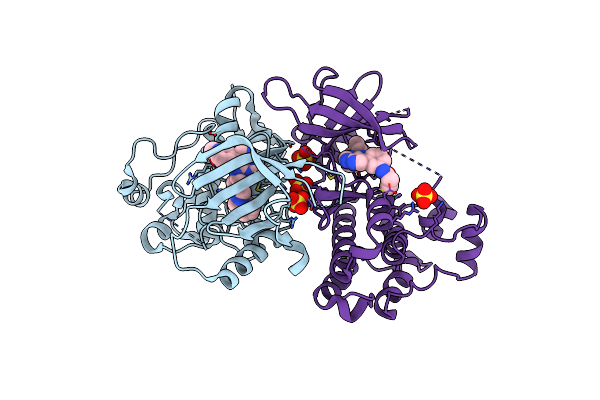 |
Crystal Structure Of Egfr T790M/V948R In Complex With Covalent Pyrrolopyrimidine 14D
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-09-30 Classification: TRANSFERASE Ligands: N7K, SO4 |
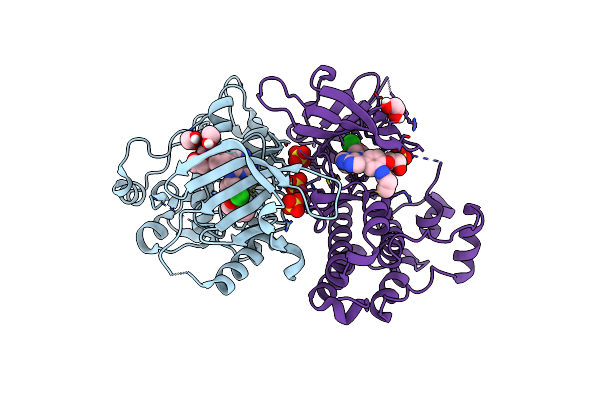 |
Crystal Structure Of Egfr T790M/V948R In Complex With Covalent Pyrrolopyrimidine 18B
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2020-09-30 Classification: TRANSFERASE Ligands: N7Q, EDO, SO4, CL |
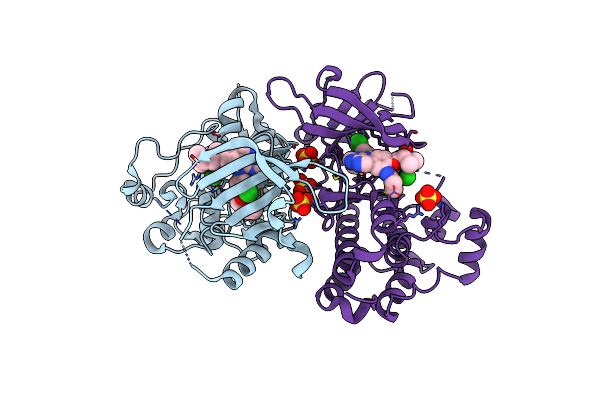 |
Crystal Structure Of Egfr T790M/V948R In Complex With Covalent Pyrrolopyrimidine 18D
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-09-30 Classification: TRANSFERASE Ligands: N7W, SO4, MG, CL |
 |
Crystal Structure Of Egfr T790M/V948R In Complex With Covalent Pyrrolopyrimidine 18C
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-09-30 Classification: TRANSFERASE Ligands: N7Z, SO4 |
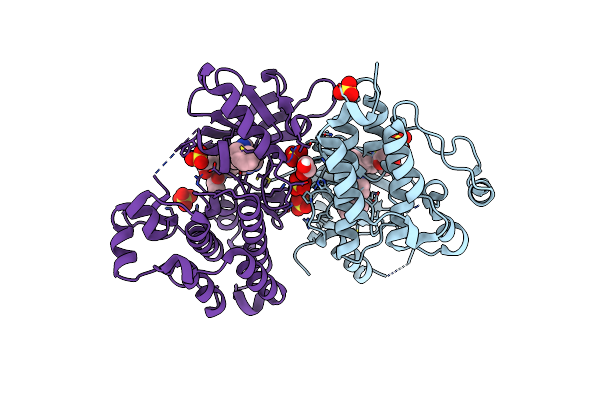 |
Crystal Structure Of Egfr T790M/V948R In Complex With Covalent Pyrrolopyrimidine 19
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-09-30 Classification: TRANSFERASE Ligands: N7B, SO4, EDO |
 |
Crystal Structure Of Egfr T790M/V948R In Complex With Covalent Pyrrolopyrimidine 21A
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2020-09-30 Classification: TRANSFERASE Ligands: N78, EDO, SO4 |
 |
Crystal Structure Of Egfr T790M/V948R In Complex With Covalent Pyrrolopyrimidine 21B
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-09-30 Classification: TRANSFERASE Ligands: N82, EDO, SO4, NA |
 |
Organism: Methanosarcina barkeri
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2014-11-05 Classification: SIGNALING PROTEIN Ligands: GDP, MG, GOL, SO4 |

