Search Count: 28
 |
Crystal Structure Of L-2-Keto-3-Deoxyfuconate 4-Dehydrogenase From Herbaspillum Huttiense (Apo Form)
Organism: Herbaspirillum huttiense
Method: X-RAY DIFFRACTION Resolution:1.27 Å Release Date: 2024-07-10 Classification: OXIDOREDUCTASE Ligands: PEG |
 |
Crystal Structure Of L-2-Keto-3-Deoxyfuconate 4-Dehydrogenase Bound To Nad(H) And Sulfate Ion
Organism: Herbaspirillum huttiense
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2024-07-10 Classification: OXIDOREDUCTASE Ligands: NAD, SO4, PEG, TRS, GOL |
 |
Crystal Structure Of L-2-Keto-3-Deoxyfuconate 4-Dehydrogenase Bound To L-Kdf Or L-2,4-Dkdf
Organism: Herbaspirillum huttiense
Method: X-RAY DIFFRACTION Resolution:1.29 Å Release Date: 2024-07-10 Classification: OXIDOREDUCTASE Ligands: TRS, A1LXA, A1LXB, PEG |
 |
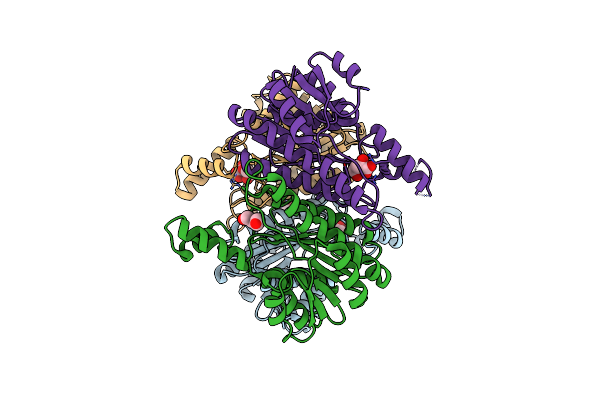
Crystal Structure Of L-2-Keto-3-Deoxyfuconate 4-Dehydrogenase Bound To L-2,4-Dkdf And Nadh
Organism: Herbaspirillum huttiense
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2024-07-10 Classification: OXIDOREDUCTASE Ligands: NAD, A1LXB |
 |
Crystal Structure Of L-2-Keto-3-Deoxyfuconate 4-Dehydrogenase Bound To D-Kdp
Organism: Herbaspirillum huttiense
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2024-07-10 Classification: OXIDOREDUCTASE Ligands: TRS, A1LXD, PEG |
 |
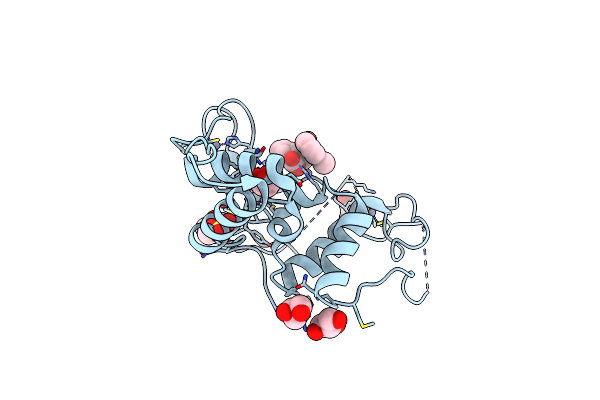
Crystal Structure Of The Ubiquitin-Like Domain In The Sf3A1 Subunit Of Human U2 Snrnp Complexed With The Stem-Loop 4 Of U1 Snrna
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-05-17 Classification: RNA BINDING PROTEIN/RNA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2020-04-15 Classification: LIGASE Ligands: ZN, D5U, GOL, EPE |
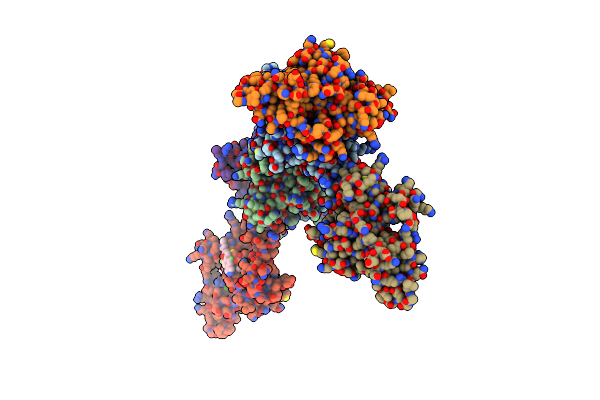 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2020-04-15 Classification: LIGASE Ligands: ZN, D60, CL, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.09 Å Release Date: 2020-01-15 Classification: SIGNALING PROTEIN |
 |
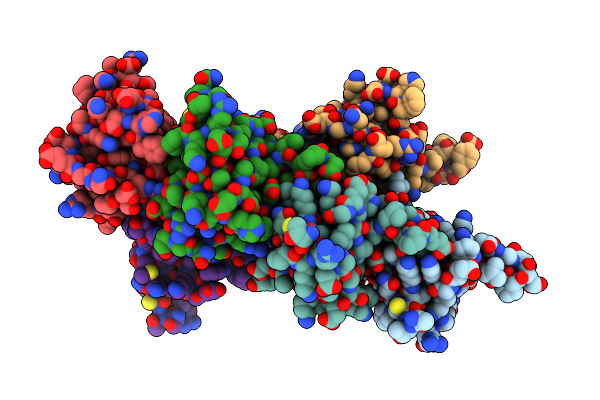
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2019-02-20 Classification: SIGNALING PROTEIN |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2017-09-06 Classification: SIGNALING PROTEIN |
 |
Organism: Danio rerio
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2017-09-06 Classification: SIGNALING PROTEIN |
 |
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2015-10-21 Classification: CELL INVASION |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2015-04-08 Classification: TRANSPORT PROTEIN Ligands: PO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.71 Å Release Date: 2015-01-14 Classification: PROTEIN BINDING |
 |
Crystal Structure Of Ethanolamine Ammonia-Lyase From Escherichia Coli Complexed With Cn-Cbl And Ethanolamine
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2010-06-02 Classification: LYASE Ligands: ETA, GOL, B12, NA |
 |
Crystal Structure Of Ethanolamine Ammonia-Lyase From Escherichia Coli Complexed With Cn-Cbl And 2-Amino-1-Propanol
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2010-06-02 Classification: LYASE Ligands: 2A1, GOL, B12 |
 |
Crystal Structure Of Ethanolamine Ammonia-Lyase From Escherichia Coli Complexed With Cn-Cbl (Substrate-Free Form)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2010-06-02 Classification: LYASE Ligands: GOL, NA, B12 |
 |
Crystal Structure Of Ethanolamine Ammonia-Lyase From Escherichia Coli Complexed With Adeninylpentylcobalamin And Ethanolamine
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2010-06-02 Classification: LYASE Ligands: COY, ETA, GOL, SO4 |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:4.50 Å Release Date: 2009-11-24 Classification: SIGNALING PROTEIN |

