Search Count: 58
 |
Crystal Structure Of Streptomyces Sp. Sulfotransferase Cpz8 Involved In Caprazamycins Synthesis
Organism: Streptomyces sp.
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: BIOSYNTHETIC PROTEIN Ligands: PO4, MG |
 |
Crystal Structure Of Streptomyces Sp. Sulfotransferase Cpz8 In Complex With Pap, Involved In Caprazamycin Synthesis
Organism: Streptomyces sp.
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: BIOSYNTHETIC PROTEIN Ligands: GOL, A3P, SO4 |
 |
Crystal Structure Of Streptomyces Sp. Sulfotransferase Cpz4 Involved In Caprazamycins Synthesis
Organism: Streptomyces sp.
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: BIOSYNTHETIC PROTEIN Ligands: FMT, MG |
 |
Crystal Structure Of The Reaction Intermediate Of Streptomyces Sp. Sulfotransferase Cpz4 Involved In Caprazamycins Synthesis
Organism: Streptomyces sp.
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: BIOSYNTHETIC PROTEIN Ligands: SO4, MG |
 |
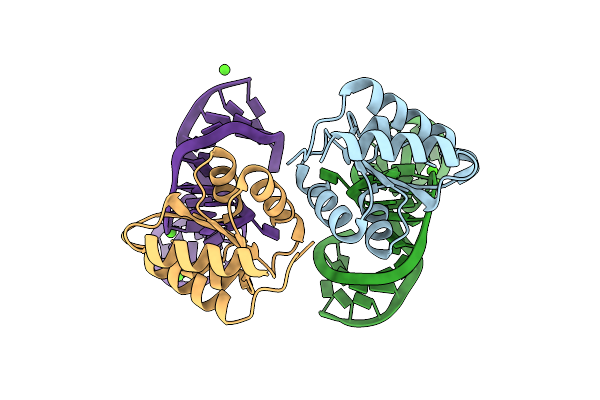
Crystal Structure Of The L7Ae Derivative Protein Ls4 In Complex With Its Co-Evolved Target Cs1 Rna
Organism: Archaeoglobus fulgidus (strain atcc 49558 / dsm 4304 / jcm 9628 / nbrc 100126 / vc-16), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2025-04-02 Classification: RNA BINDING PROTEIN Ligands: MG |
 |
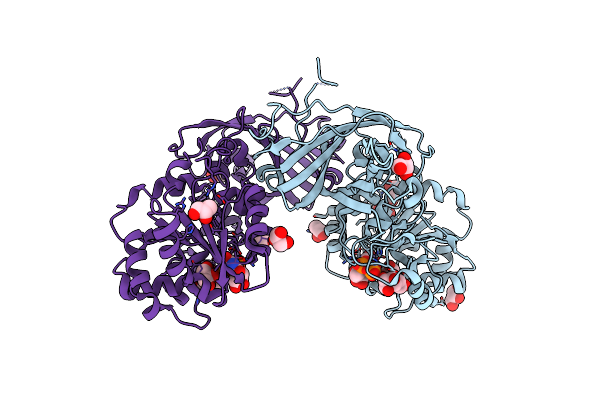
Crystal Structure Of The L7Ae Derivative Protein Ls12 In Complex With Its Co-Evolved Target Cs2 Rna
Organism: Archaeoglobus fulgidus (strain atcc 49558 / dsm 4304 / jcm 9628 / nbrc 100126 / vc-16), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.12 Å Release Date: 2025-04-02 Classification: RNA BINDING PROTEIN Ligands: CA |
 |
Organism: Archaeoglobus fulgidus dsm 4304
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-04-02 Classification: RNA BINDING PROTEIN |
 |
Crystal Structure Of Aspergillus Fumigatus Galactofuranosylransferase (Afgfsa) In Complex With Udp And Galactofuranose
Organism: Aspergillus fumigatus cea10
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2024-10-30 Classification: TRANSFERASE Ligands: MN, UDP, GOL, GZL |
 |
Cryo-Em Structure Of Aquifex Aeolicus Minimal Protein-Only Rnase P (Harp) In Complex With Pre-Trnas
Organism: Aquifex aeolicus vf5
Method: ELECTRON MICROSCOPY Release Date: 2024-08-14 Classification: HYDROLASE-RNA COMPLEX |
 |
Cryo-Em Structure Of Hydrogenobacter Thermophilus Minimal Protein-Only Rnase P (Harp) In Complex With Pre-Trnas
Organism: Hydrogenobacter thermophilus dsm 653, Aquifex aeolicus
Method: ELECTRON MICROSCOPY Release Date: 2024-08-14 Classification: HYDROLASE/RNA Ligands: MG |
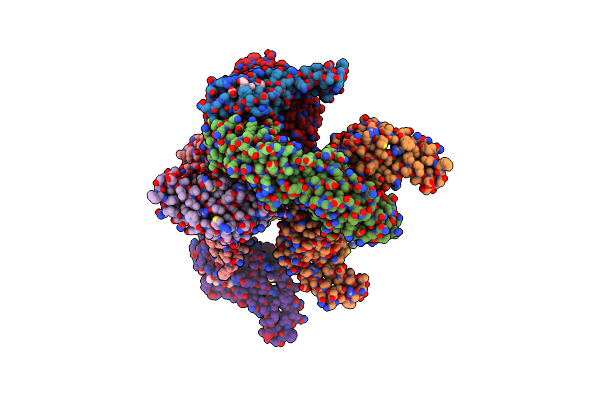 |
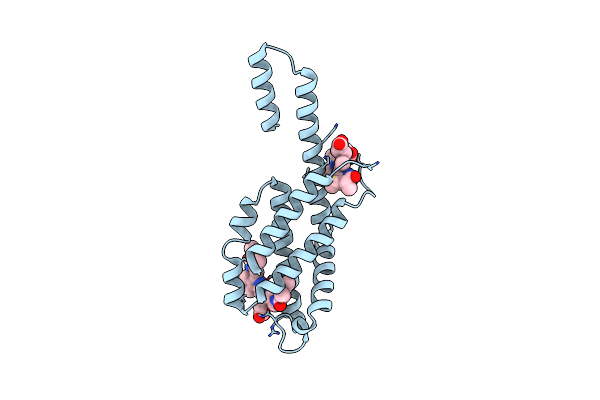
Crystal Structure Of The Dissociated C-Phycocyanin Alpha-Chain From Thermoleptolyngbya Sp. O-77
Organism: Leptolyngbya sp. o-77
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-05-22 Classification: PHOTOSYNTHESIS Ligands: CYC, MG |
 |
Crystal Structure Of The Dissociated C-Phycocyanin Beta-Chain From Thermoleptolyngbya Sp. O-77
Organism: Leptolyngbya sp. o-77
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2024-05-22 Classification: PHOTOSYNTHESIS Ligands: CYC |
 |
Crystal Structure Of The Reassembled C-Phycocyanin Hexamer From Thermoleptolyngbya Sp. O-77
Organism: Leptolyngbya sp. o-77
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2024-05-22 Classification: PHOTOSYNTHESIS Ligands: CYC, NA, PO4 |
 |
Crystal Structure Of The Reassembled C-Phycocyanin Octamer From Thermoleptolyngbya Sp. O-77
Organism: Leptolyngbya sp. o-77
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-05-22 Classification: PHOTOSYNTHESIS Ligands: CYC, PO4 |
 |
Organism: Micromonospora sp.
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2024-05-15 Classification: TRANSFERASE Ligands: GOL, A3P |
 |
Crystal Structure Of Tick Tyrosylprotein Sulfotransferase Reveals The Activation Mechanism Of Tick Anticoagulant Protein Madanin
Organism: Ixodes, Longicornis species complex
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2024-04-10 Classification: TRANSFERASE Ligands: A3P, 1PS, TRS, MG |
 |
Crystal Structure Of Anti-Peg Antibody M9 Fv-Clasp Fragment With Peg (Co-Crystallization With Peg3350)
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2024-03-20 Classification: IMMUNE SYSTEM Ligands: 12P |
 |
Crystal Structure Of Anti-Peg Antibody M9 Fv-Clasp Fragment With Peg (Co-Crystallization With Peg2000Mme)
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2024-03-20 Classification: IMMUNE SYSTEM Ligands: 12P |
 |
Crystal Structure Of Anti-Peg Antibody M9 Fv-Clasp Fragment With Peg (Co-Crystallization With Peg550Dme)
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2024-03-20 Classification: IMMUNE SYSTEM Ligands: HZA |
 |
Crystal Structure Of Anti-Peg Antibody M11 Fv-Clasp Fragment With Peg (Co-Crystallization With Peg3350)
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2024-03-20 Classification: IMMUNE SYSTEM Ligands: SO4, PE8 |

