Search Count: 17
 |
Organism: Enterococcus faecalis
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2024-02-28 Classification: HYDROLASE |
 |
Organism: Gluconobacter oxydans
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2023-11-08 Classification: TRANSFERASE Ligands: CL, NA |
 |
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-06-07 Classification: MEMBRANE PROTEIN Ligands: DAR |
 |
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-06-07 Classification: MEMBRANE PROTEIN Ligands: DLY |
 |
Organism: Enterococcus faecalis
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2022-11-30 Classification: CELL ADHESION Ligands: MG |
 |
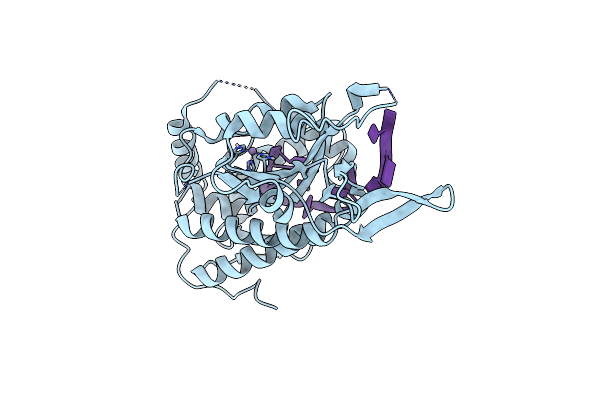
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-07-13 Classification: DNA BINDING PROTEIN Ligands: GOL, CL, MN |
 |
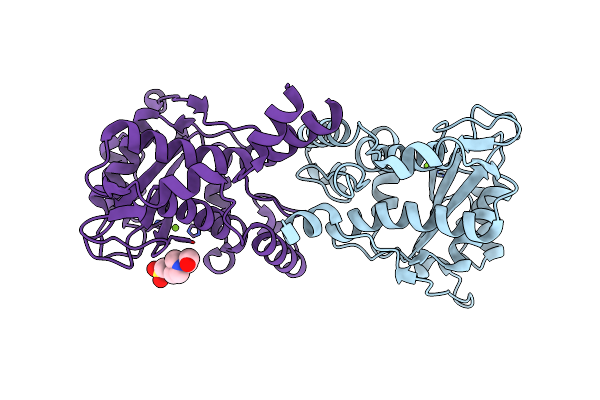
Organism: Escherichia coli, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-07-13 Classification: DNA BINDING PROTEIN Ligands: MN |
 |
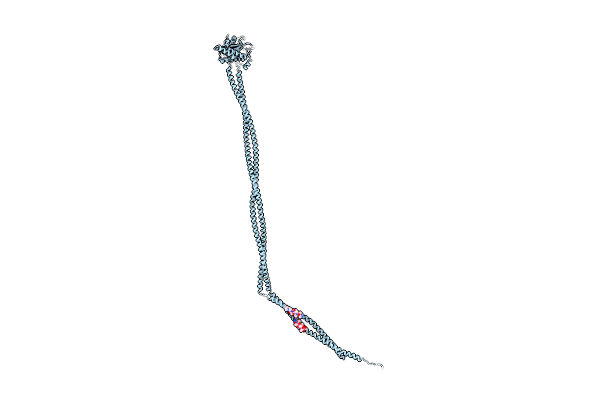
Organism: Enterococcus faecalis
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2020-09-16 Classification: CELL ADHESION Ligands: MG, EPE |
 |
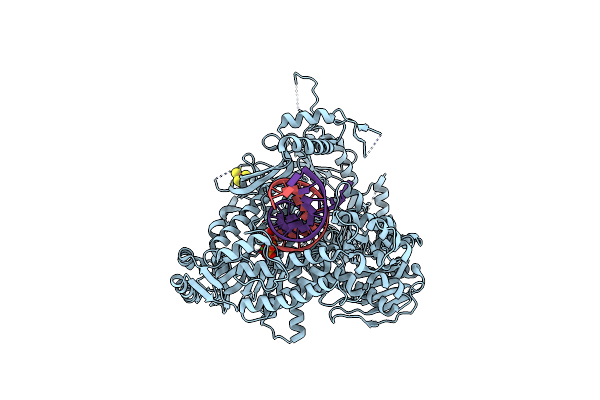
Organism: Enterococcus faecalis, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.06 Å Release Date: 2020-09-16 Classification: CELL ADHESION Ligands: SO4 |
 |
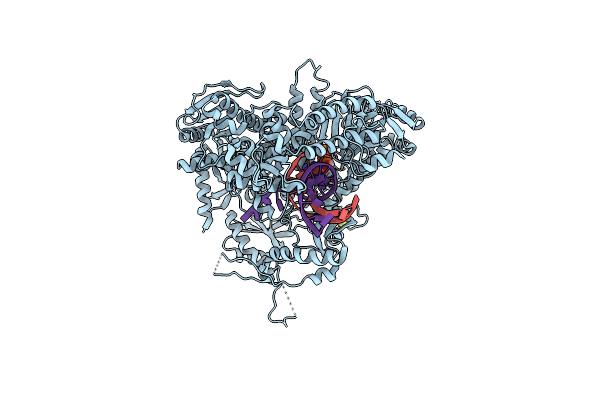
The Crystal Structure Of Pol2Core In Complex With Dna And An Incoming Nucleotide, Carrying An Fe-S Cluster
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2019-04-24 Classification: DNA BINDING PROTEIN Ligands: SF4, DTP, MG |
 |
The Crystal Structure Of Pol2Core In Complex With Dna And An Incoming Nucleotide, Carrying An Fe-S Cluster
Organism: Saccharomyces cerevisiae, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2019-04-17 Classification: DNA BINDING PROTEIN Ligands: SF4, DTP, CA |
 |
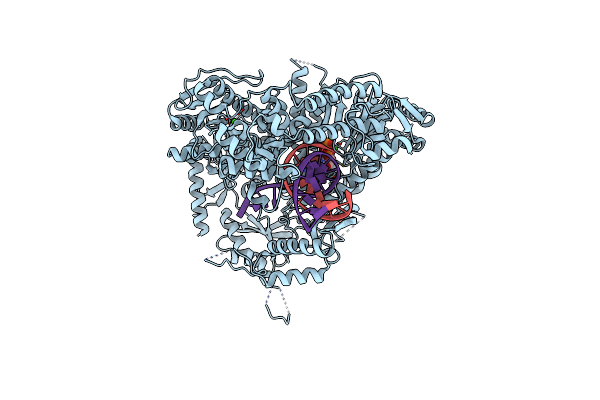
The Crystal Structure Of Pol2Core-M644G In Complex With Dna And An Incoming Nucleotide
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-01-30 Classification: DNA BINDING PROTEIN Ligands: DTP, CA, FE |
 |
The Crystal Structure Of The Pol2 Catalytic Domain Of Dna Polymerase Epsilon Carrying A P301R Substitution.
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.62 Å Release Date: 2019-01-30 Classification: DNA BINDING PROTEIN Ligands: DTP, CA, FE |
 |
The Crystal Structure Of The Pol2 Catalytic Domain Of Dna Polymerase Epsilon Carrying A P301R Substitution.
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2019-01-30 Classification: DNA BINDING PROTEIN Ligands: DTP, CA, FE |
 |
Organism: Lactobacillus delbrueckii
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2018-07-25 Classification: TRANSPORT PROTEIN |
 |
Crystal Structure At 2.1 A Of The S-Component For Biotin From An Ecf-Type Abc Transporter
Organism: Lactococcus lactis subsp. cremoris
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2012-08-29 Classification: TRANSPORT PROTEIN Ligands: BTN, BNG |
 |
Crystal Structure At 2.0 A Of The S-Component For Thiamin From An Ecf-Type Abc Transporter
Organism: Lactococcus lactis subsp. cremoris
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-06-29 Classification: THIAMINE-BINDING PROTEIN Ligands: BNG, VIB |

