Search Count: 32
 |
Organism: Burkholderia pseudomallei k96243
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2023-08-16 Classification: HYDROLASE |
 |
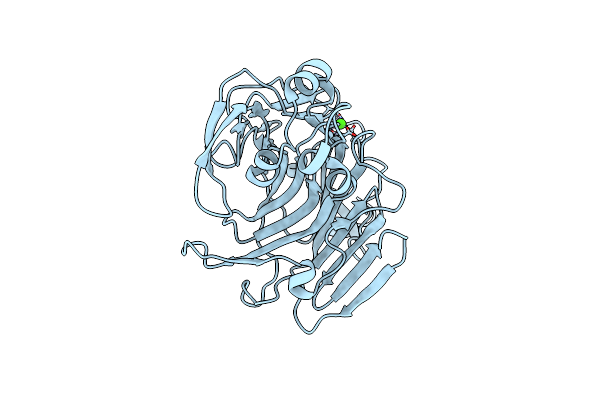
Crystal Structure Of A Selenomethionine-Labeled Bpsl1038 From Burkholderia Pseudomallei
Organism: Burkholderia pseudomallei k96243
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2023-08-16 Classification: HYDROLASE Ligands: BME, NA |
 |
Organism: Persicobacter sp. ccb-qb2
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-12-30 Classification: SUGAR BINDING PROTEIN Ligands: CA |
 |
Organism: Persicobacter sp. ccb-qb2
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2020-12-30 Classification: SUGAR BINDING PROTEIN Ligands: CA, PG4, NA |
 |
Organism: Persicobacter
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2019-08-28 Classification: LYASE Ligands: CA |
 |
Organism: Dermatophagoides farinae
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2019-03-13 Classification: ALLERGEN Ligands: 1PE, GOL, BME |
 |
Organism: Dermatophagoides farinae
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2019-03-13 Classification: ALLERGEN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-08-01 Classification: UNKNOWN FUNCTION Ligands: GOL, CL, ZN, BME |
 |
Organism: Persicobacter sp. ccb-qb2
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-06-13 Classification: LYASE Ligands: CA, K, ACT, PO4 |
 |
Organism: Methylacidiphilum infernorum
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2014-09-24 Classification: OXYGEN BINDING Ligands: HEM, PO4 |
 |
Organism: Methylacidiphilum infernorum
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2014-09-24 Classification: OXYGEN BINDING Ligands: HEM |
 |
Organism: Methylacidiphilum infernorum
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2014-08-06 Classification: OXYGEN TRANSPORT Ligands: HEM, MPD |
 |
Organism: Methylacidiphilum infernorum
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2014-08-06 Classification: OXYGEN TRANSPORT Ligands: HEM, HEZ, IMD |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2014-01-22 Classification: HYDROLASE Ligands: MG |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2014-01-22 Classification: HYDROLASE Ligands: MG |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.06 Å Release Date: 2014-01-22 Classification: HYDROLASE Ligands: MN |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.22 Å Release Date: 2014-01-22 Classification: HYDROLASE Ligands: MN |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-01-22 Classification: HYDROLASE Ligands: MN, PEG |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2014-01-22 Classification: HYDROLASE Ligands: CO |
 |
Crystal Structure Of Alpha-Amylase From Geobacillus Thermoleovorans, Gta, Complexed With Acarbose
Organism: Geobacillus thermoleovorans
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2013-03-13 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: CA, ACI |

