Search Count: 31
 |
Organism: Uncultured organism
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2016-10-05 Classification: HYDROLASE |
 |
Crystal Structure Of Mgs-Mt1, An Alpha/Beta Hydrolase Enzyme From A Lake Matapan Deep-Sea Metagenome Library
Organism: Unidentified
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2015-03-04 Classification: HYDROLASE Ligands: CL, MES, GOL |
 |
Crystal Structure Of Mgs-M1, An Alpha/Beta Hydrolase Enzyme From A Medee Basin Deep-Sea Metagenome Library
Organism: Unidentified
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2015-02-25 Classification: HYDROLASE Ligands: CL, F, PGE |
 |
Crystal Structure Of Mgs-M2, An Alpha/Beta Hydrolase Enzyme From A Medee Basin Deep-Sea Metagenome Library
Organism: Unidentified
Method: X-RAY DIFFRACTION Resolution:3.01 Å Release Date: 2015-02-25 Classification: HYDROLASE Ligands: GOL |
 |
Crystal Structure Of Mgs-M4, An Aldo-Keto Reductase Enzyme From A Medee Basin Deep-Sea Metagenome Library
Organism: Unidentified
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2015-02-25 Classification: HYDROLASE Ligands: SO4, NA |
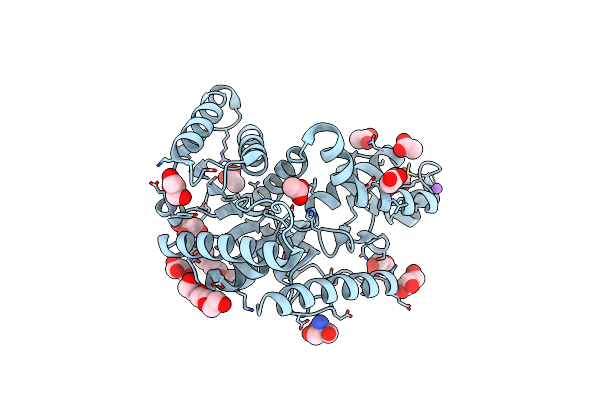 |
Crystal Structure Of Mgs-M5, A Lactate Dehydrogenase Enzyme From A Medee Basin Deep-Sea Metagenome Library
Organism: Unidentified
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2015-02-25 Classification: HYDROLASE |
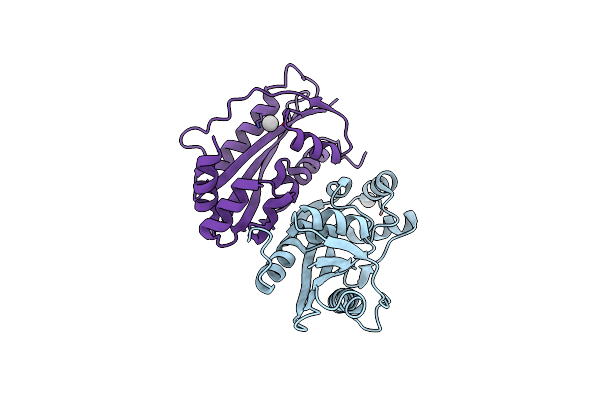 |
Crystal Structure Of The Uncharacterized Maf Protein Ycef From E. Coli, Mutant D69A
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2013-08-14 Classification: HYDROLASE Ligands: UNX |
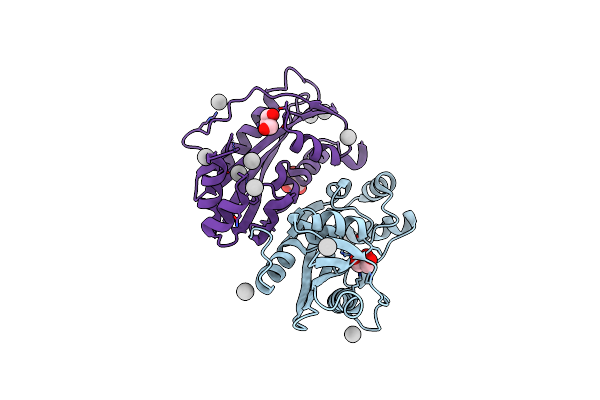 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2013-03-20 Classification: CELL CYCLE Ligands: GOL, UNX |
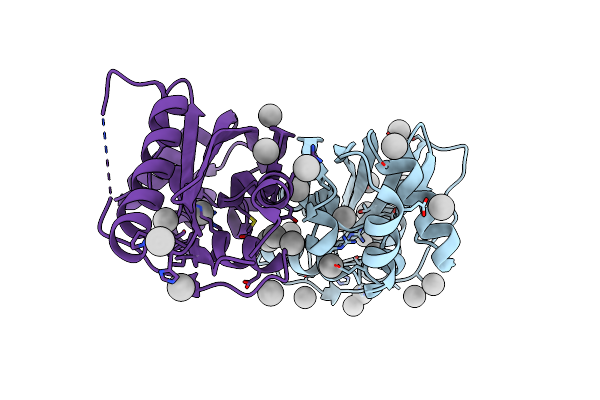 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2012-10-24 Classification: CELL CYCLE Ligands: UNX |
 |
Crystal Structure Of A Putative Fumarylacetoacetate Isomerase/Hydrolase From Oleispira Antarctica
Organism: Oleispira antarctica
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2012-01-18 Classification: HYDROLASE Ligands: ZN, TAR, ACT |
 |
Crystal Structure Of A Putative Kdpg (2-Keto-3-Deoxy-6-Phosphogluconate) Aldolase From Oleispira Antarctica
Organism: Oleispira antarctica
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2012-01-18 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION Ligands: PYR |
 |
Bromide Soaked Structure Of An Esterase From The Oil-Degrading Bacterium Oleispira Antarctica
Organism: Oleispira antarctica
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2011-06-15 Classification: HYDROLASE Ligands: BR |
 |
Organism: Oleispira antarctica
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-04-13 Classification: HYDROLASE Ligands: SO4, NA, CL, CA |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2011-03-16 Classification: HYDROLASE Ligands: CL |
 |
Crystal Structure Of A Serine Dehydrogenase From Pseudomonas Aeruginosa Pao1 In Complex With Nad
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2011-02-23 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2010-10-27 Classification: HYDROLASE Ligands: DUP, MG |
 |
Crystal Structure Of A Possible 3-Hydroxyisobutyrate Dehydrogenase From Pseudomonas Aeruginosa Pao1
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2010-08-18 Classification: OXIDOREDUCTASE Ligands: ACT, EDO, EPE |
 |
Organism: Oleispira antarctica
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2010-06-23 Classification: TRANSFERASE |
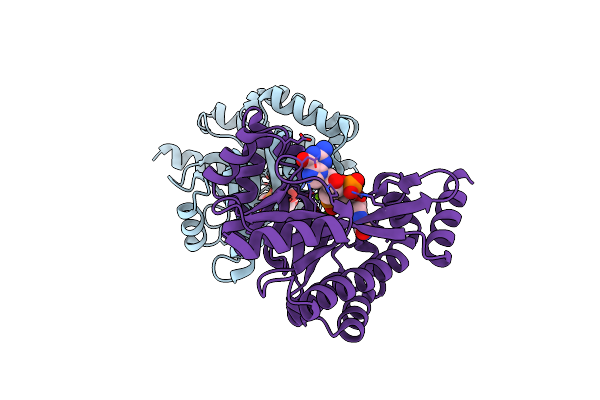 |
Crystal Structure Of Putative Diguanylate Cyclase/Phosphodiesterase Complex With Cyclic Di-Gmp
Organism: Thiobacillus denitrificans
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2010-06-16 Classification: Structural Genomics, Unknown function Ligands: C2E, MG, CL |
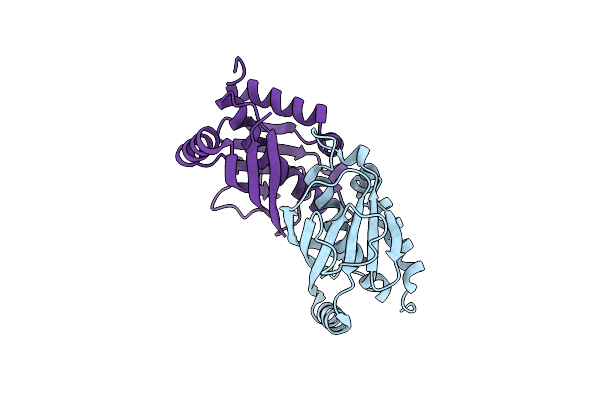 |
The Crystal Structure Of The Protein Olei01261 With Unknown Function From Chlorobaculum Tepidum Tls
Organism: Oleispira antarctica rb-8
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2010-03-16 Classification: Structural Genomics, Unknown function |

