Search Count: 28
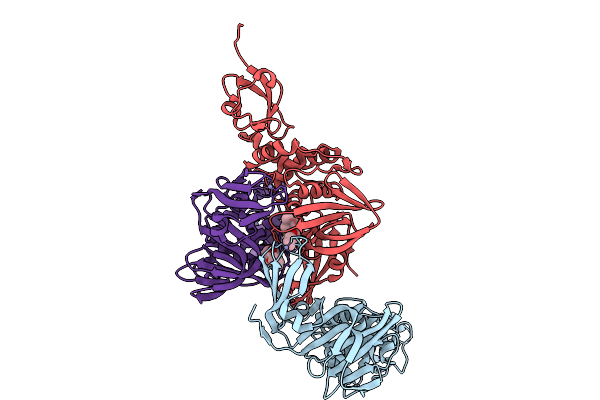 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2026-01-21 Classification: VIRAL PROTEIN Ligands: A1CLE, ZN |
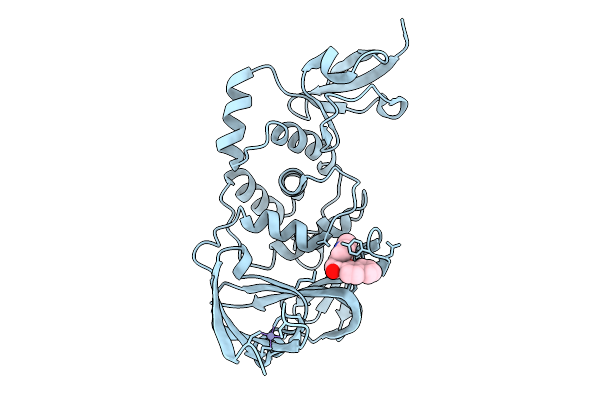 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2026-01-21 Classification: VIRAL PROTEIN Ligands: ZN, A1CLF |
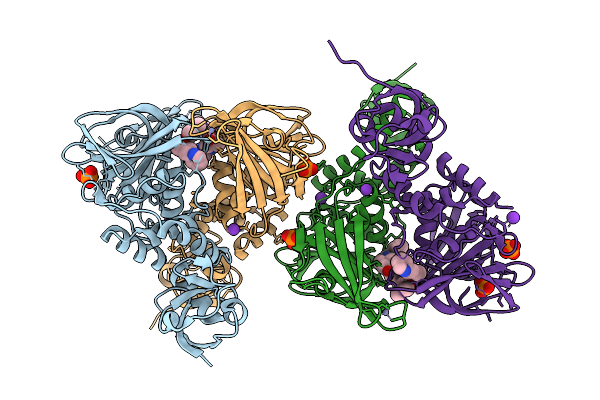 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2026-01-21 Classification: VIRAL PROTEIN Ligands: ZN, A1CLD, PO4, K |
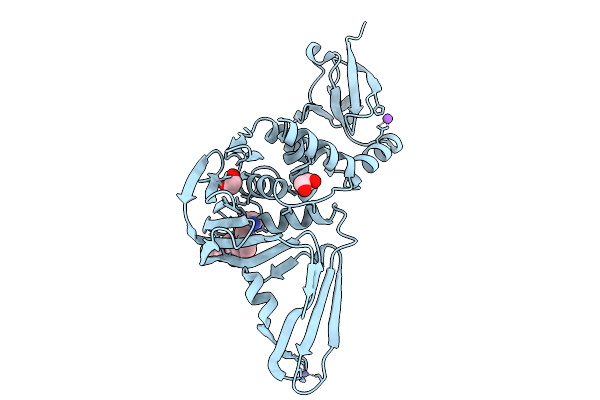 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2026-01-21 Classification: VIRAL PROTEIN Ligands: ZN, EDO, A1CLN, NA |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2026-01-21 Classification: VIRAL PROTEIN Ligands: ZN, A1CLP, GOL |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2026-01-21 Classification: VIRAL PROTEIN Ligands: ZN, A1CLO, EDO |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2026-01-21 Classification: VIRAL PROTEIN Ligands: ZN, EDO, A1CLL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.93 Å Release Date: 2025-11-26 Classification: LIGASE Ligands: A1CI7, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2025-11-26 Classification: LIGASE Ligands: A1CI8, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2025-11-26 Classification: LIGASE Ligands: A1CI9 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2025-11-26 Classification: LIGASE Ligands: A1CLY |
 |
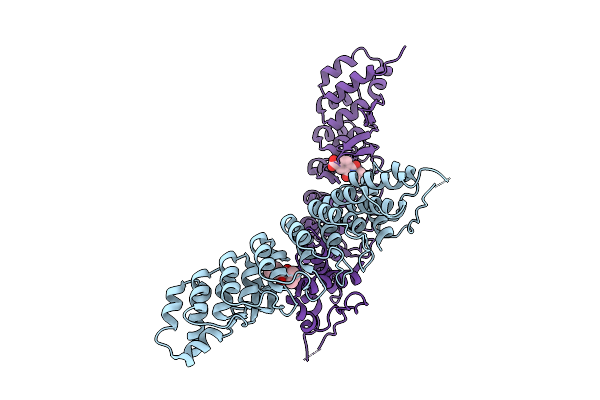
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2025-11-26 Classification: LIGASE Ligands: A1CL1, SO4 |
 |
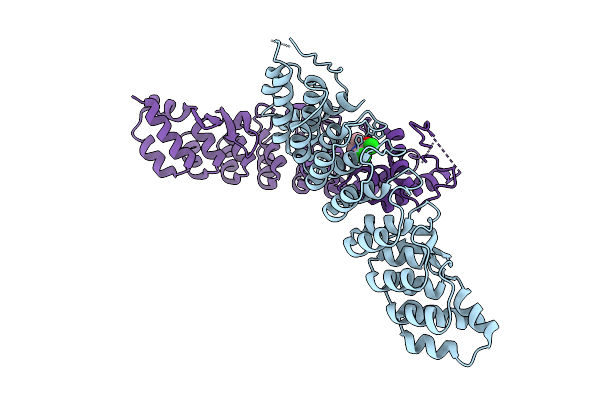
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.05 Å Release Date: 2025-11-26 Classification: LIGASE Ligands: A1CMX |
 |
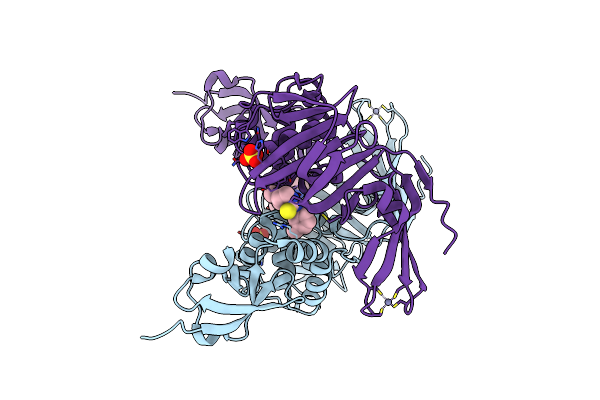
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2025-11-26 Classification: LIGASE Ligands: A1CMY |
 |
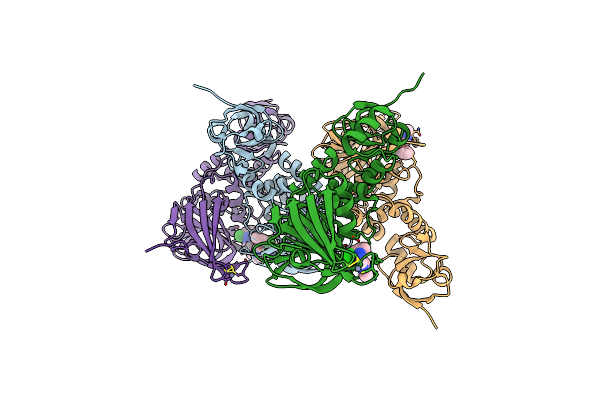
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2024-07-31 Classification: VIRAL PROTEIN, HYDROLASE Ligands: A1ASK, ZN, SO4, CL |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-07-31 Classification: VIRAL PROTEIN, HYDROLASE Ligands: A1ARL, ZN, CL |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-07-31 Classification: VIRAL PROTEIN, HYDROLASE Ligands: A1ASL, ZN, SO4 |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-12-07 Classification: OXIDOREDUCTASE Ligands: CU, 010 |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2022-12-07 Classification: OXIDOREDUCTASE Ligands: IMD |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-12-07 Classification: OXIDOREDUCTASE Ligands: BYZ, PGE |

