Search Count: 8
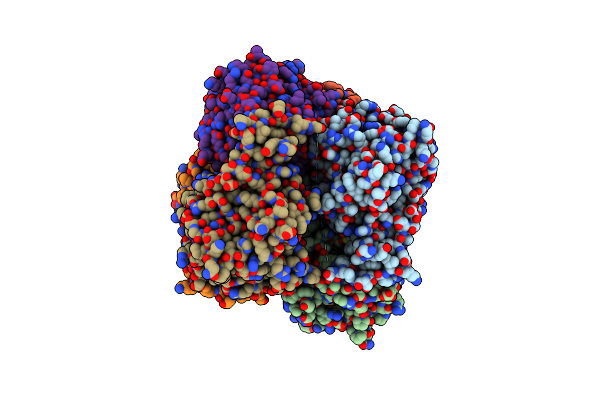 |
Crystal Structure Of Metformin Hydrolase (Mfmab) From Pseudomonas Mendocina Sp. Met-2 With Ni2+2 Bound
Organism: Pseudomonas mendocina
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-03-06 Classification: HYDROLASE Ligands: NI |
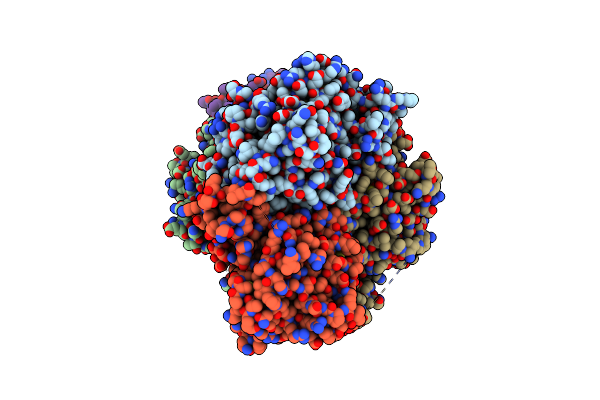 |
Crystal Structure Of Metformin Hydrolase (Mfmab) From Pseudomonas Mendocina Sp. Met-2 Mutant (Mfma/D188N)
Organism: Pseudomonas mendocina
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-03-06 Classification: HYDROLASE Ligands: ZN |
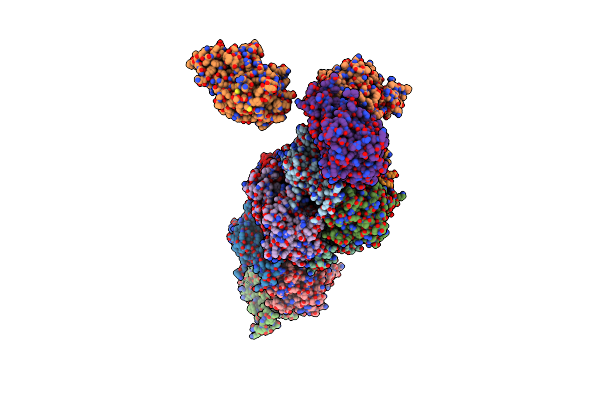 |
Crystal Structure Of Metformin Hydrolase (Mfmab) From Pseudomonas Mendocina Sp. Met-2 Apo Form
Organism: Pseudomonas mendocina
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-03-06 Classification: HYDROLASE Ligands: NI |
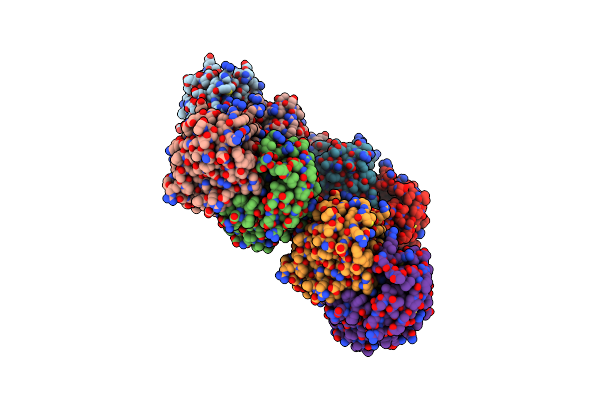 |
Organism: Herbaspirillum sp. bh-1
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-11-18 Classification: HYDROLASE Ligands: BME |
 |
Triuret Hydrolase (Trta) From Herbaspirillum Sp. Bh-1 C162S Bound With Biuret
Organism: Herbaspirillum sp. bh-1
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2020-11-18 Classification: HYDROLASE Ligands: EDO, C5J |
 |
Triuret Hydrolase (Trta) From Herbaspirillum Sp. Bh-1 C162S Bound With Triuret
Organism: Herbaspirillum sp. bh-1
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2020-11-18 Classification: HYDROLASE Ligands: TIU, EDO |
 |
Organism: Rhodococcus sp. mel
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2020-11-18 Classification: HYDROLASE Ligands: C5J |
 |
Organism: Rhodococcus sp. mel
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-11-18 Classification: HYDROLASE |

