Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
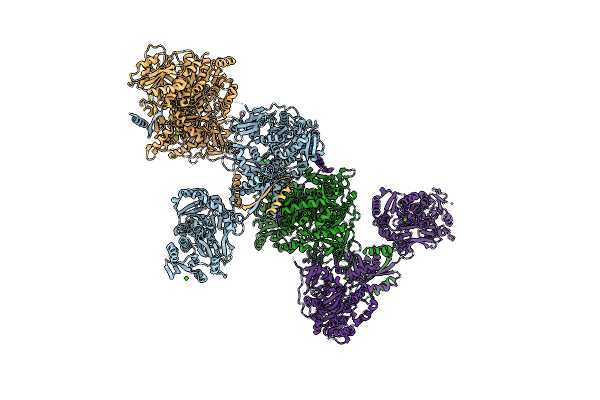 |
Adenylation, Ketoreductase, And Pseudo Asub Multidomain Structure Of A Keto Acid-Selecting Nrps Module
Organism: Bacillus stratosphericus lama 585
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2020-02-19 Classification: BIOSYNTHETIC PROTEIN Ligands: CA, MG |
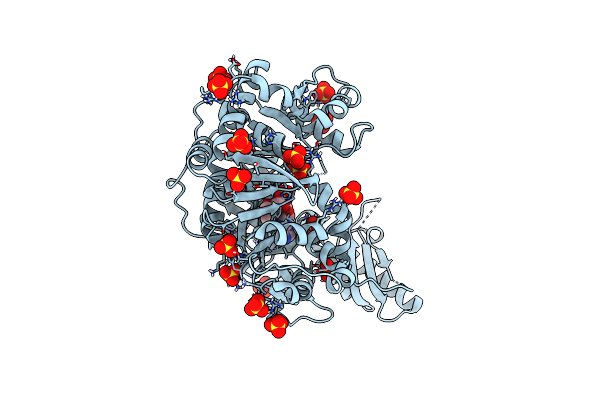 |
Adenylation Domain Of A Keto Acid-Selecting Nrps Module Bound To Keto Acyl Adenylate Space Group P43212
Organism: Bacillus stratosphericus lama 585
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2020-02-19 Classification: BIOSYNTHETIC PROTEIN Ligands: QA7, SO4 |
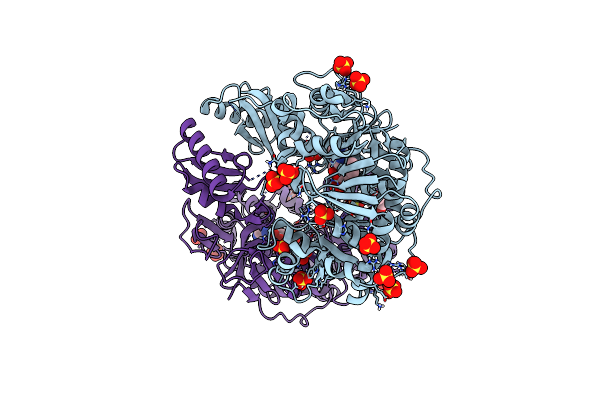 |
Adenylation Domain Of A Keto Acid-Selecting Nrps Module Bound To Keto Acyl Adenylate Space Group P212121
Organism: Bacillus stratosphericus lama 585
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2020-02-19 Classification: BIOSYNTHETIC PROTEIN Ligands: QA7, SO4, GOL |
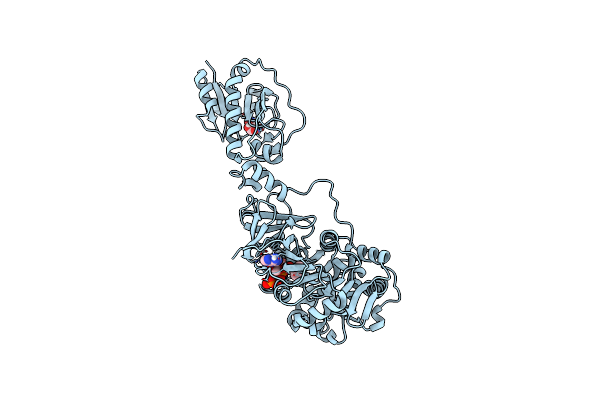 |
Organism: Brevibacillus parabrevis
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2020-02-19 Classification: BIOSYNTHETIC PROTEIN Ligands: FMT, APC, KIV |
 |
Organism: Caulobacter vibrioides (strain atcc 19089 / cb15)
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2019-11-27 Classification: DNA BINDING PROTEIN |
 |
Organism: Caulobacter vibrioides (strain atcc 19089 / cb15)
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2019-11-27 Classification: DNA BINDING PROTEIN Ligands: CD |
 |
Organism: Caulobacter vibrioides (strain atcc 19089 / cb15)
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2019-11-27 Classification: DNA BINDING PROTEIN |
 |
Organism: Geobacillus sp.
Method: X-RAY DIFFRACTION Resolution:3.08 Å Release Date: 2017-05-10 Classification: HYDROLASE/INHIBITOR Ligands: MJ8 |
 |
Organism: Escherichia coli, Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:5.90 Å Release Date: 2014-07-09 Classification: RIBOSOME |
 |
Organism: Escherichia coli, Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:6.76 Å Release Date: 2014-07-09 Classification: RIBOSOME |
 |
Crystal Structure Of The Whole Ribosomal Complex With A Stop Codon In The A-Site.
Organism: Escherichia coli, Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:6.46 Å Release Date: 2014-07-09 Classification: RIBOSOME |
 |
Structure Of The Tatc Core Of The Twin Arginine Protein Translocation System
Organism: Aquifex aeolicus
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2012-12-05 Classification: TRANSPORT PROTEIN Ligands: LMN |
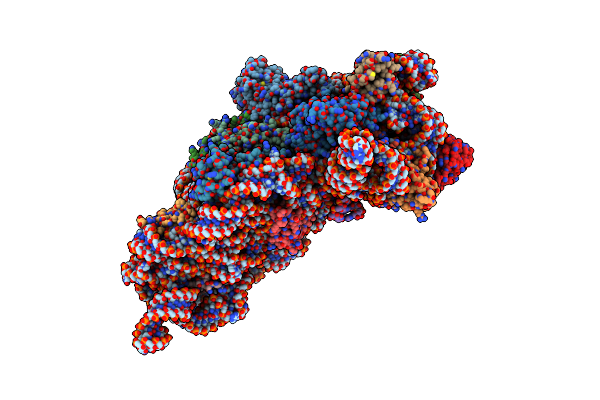 |
Structure Of The Thermus Thermophilus 30S Ribosomal Subunit Bound To Codon And Near-Cognate Transfer Rna Anticodon Stem-Loop Mismatched At The First Codon Position At The A Site With Paromomycin
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2002-11-29 Classification: RIBOSOME Ligands: PAR, MG, ZN |
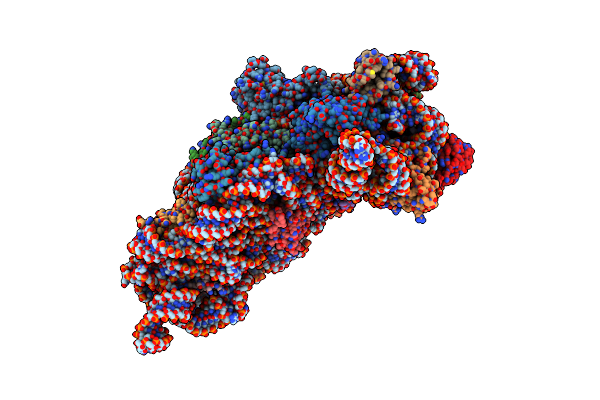 |
Structure Of The Thermus Thermophilus 30S Ribosomal Subunit Bound To Codon And Near-Cognate Transfer Rna Anticodon Stem-Loop Mismatched At The Second Codon Position At The A Site With Paromomycin
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:3.35 Å Release Date: 2002-11-29 Classification: RIBOSOME Ligands: PAR, MG, ZN |
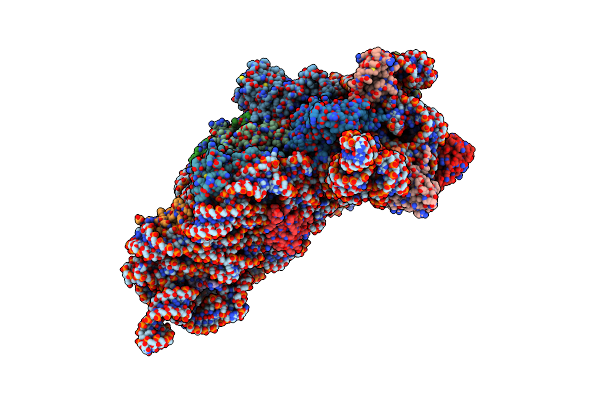 |
Structure Of The Thermus Thermophilus 30S Ribosomal Subunit In The Presence Of Codon And Crystallographically Disordered Near-Cognate Transfer Rna Anticodon Stem-Loop Mismatched At The First Codon Position
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:3.80 Å Release Date: 2002-11-29 Classification: RIBOSOME Ligands: ZN |
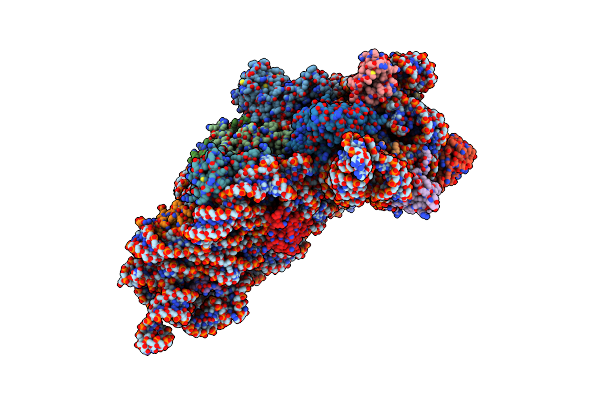 |
Structure Of The Thermus Thermophilus 30S Ribosomal Subunit In The Presence Of Crystallographically Disordered Codon And Near-Cognate Transfer Rna Anticodon Stem-Loop Mismatched At The Second Codon Position
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:3.65 Å Release Date: 2002-11-29 Classification: RIBOSOME Ligands: ZN |
 |
Structure Of The Thermus Thermophilus 30S Ribosomal Subunit In Complex With The Antibiotic Paromomycin
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:3.31 Å Release Date: 2001-05-04 Classification: RIBOSOME Ligands: PAR, MG, ZN |
 |
Structure Of The Thermus Thermophilus 30S Ribosomal Subunit In Complex With A Messenger Rna Fragment And Cognate Transfer Rna Anticodon Stem-Loop Bound At The A Site And With The Antibiotic Paromomycin
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:3.11 Å Release Date: 2001-05-04 Classification: RIBOSOME Ligands: PAR, MG, ZN |
 |
Structure Of The Thermus Thermophilus 30S Ribosomal Subunit In Complex With A Messenger Rna Fragment And Cognate Transfer Rna Anticodon Stem-Loop Bound At The A Site
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:3.31 Å Release Date: 2001-05-04 Classification: RIBOSOME Ligands: MG, ZN |