Search Count: 15
 |
Organism: Neisseria meningitidis
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: VIRUS |
 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: CELL ADHESION |
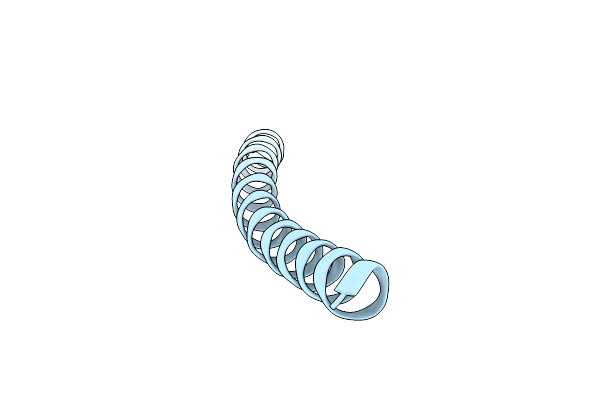 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-12-20 Classification: VIRUS |
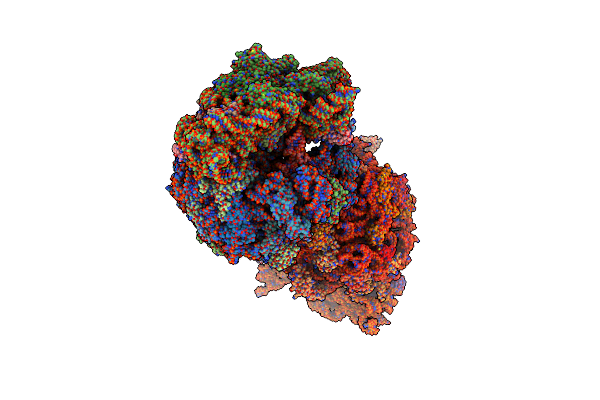 |
Crystal Structure Of The Thermus Thermophilus 70S Ribosome In Complex With Amikacin, Mrna, And A-, P-, And E-Site Trnas
Organism: Escherichia coli, Escherichia phage t4, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2023-08-09 Classification: RIBOSOME Ligands: MG, AKN, ZN, SF4 |
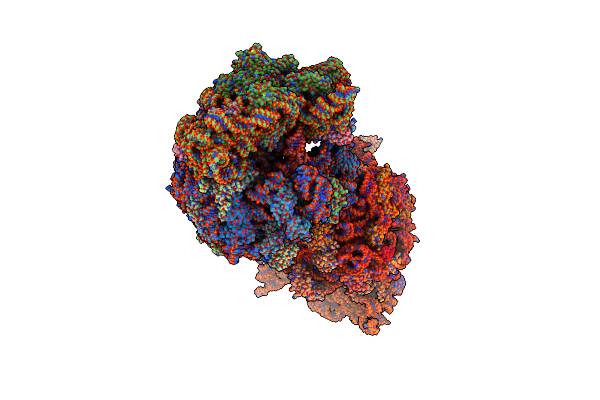 |
Crystal Structure Of The Thermus Thermophilus 70S Ribosome In Complex With Kanamycin, Mrna, And A-, P-, And E-Site Trnas
Organism: Escherichia coli, Escherichia phage t4, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2023-08-09 Classification: RIBOSOME Ligands: MG, KAN, ZN, SF4 |
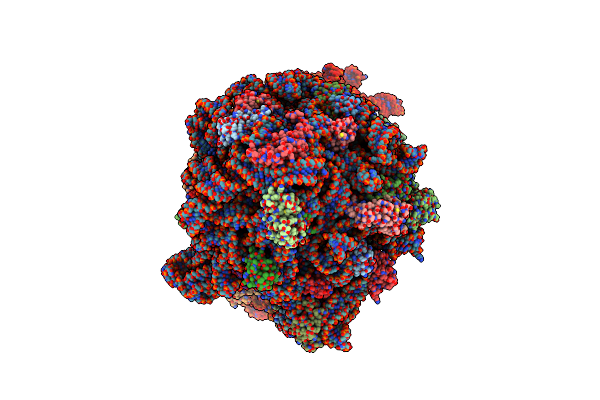 |
Cryo-Em Structure Of The Escherichia Coli 70S Ribosome In Complex With Amikacin, Mrna, And A-, P-, And E-Site Trnas
Organism: Escherichia phage t4, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-08-09 Classification: RIBOSOME Ligands: MG, AKN, PHE, ZN |
 |
Organism: Pseudomonas aeruginosa pao1
Method: ELECTRON MICROSCOPY Release Date: 2023-03-22 Classification: PROTEIN FIBRIL |
 |
Cryo-Em Structure Of Pf4 Bacteriophage Coat Protein With Single-Stranded Dna
Organism: Pseudomonas virus pf1, Pseudomonas aeruginosa pao1
Method: ELECTRON MICROSCOPY Release Date: 2020-02-26 Classification: VIRUS |
 |
Organism: Pseudomonas virus pf1
Method: ELECTRON MICROSCOPY Release Date: 2020-02-26 Classification: VIRUS |
 |
Crystal Structure Of An Ancient Sequence-Reconstructed Elongation Factor Tu (Node 262)
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-05-22 Classification: TRANSLATION Ligands: GDP, MG |
 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2016-06-15 Classification: HYDROLASE |
 |
Organism: Chaetomium thermophilum
Method: X-RAY DIFFRACTION Resolution:2.76 Å Release Date: 2016-06-15 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2016-06-15 Classification: HYDROLASE |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:10.90 Å Release Date: 2015-05-13 Classification: SIGNALING PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:10.30 Å Release Date: 2015-05-13 Classification: SIGNALING PROTEIN |

