Search Count: 20
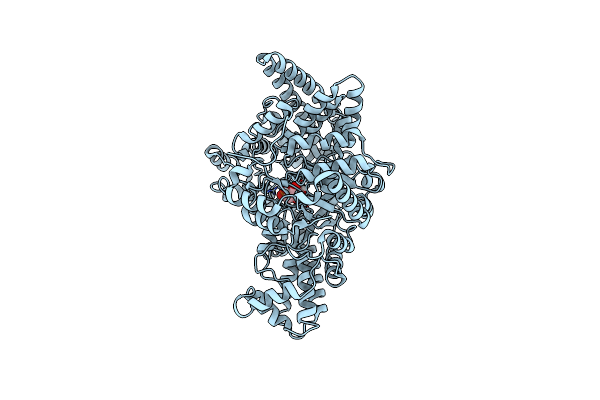 |
Crystal Structure Of Pfld Bound To 1,5-Anhydromannitol-6-Phosphate In Streptococcus Dysgalactiae Subsp. Equisimilis
Organism: Streptococcus dysgalactiae subsp. equisimilis
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2024-10-02 Classification: LYASE Ligands: 7P6 |
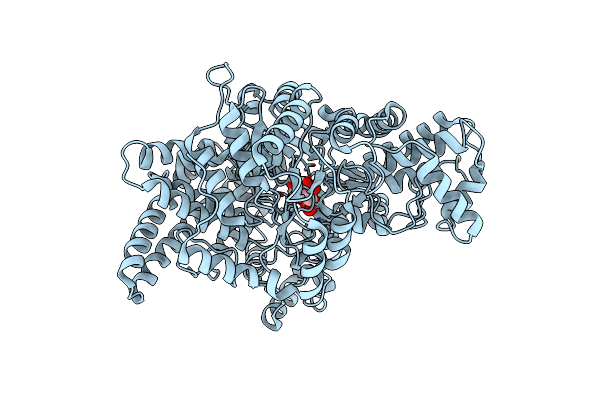 |
Crystal Structure Of Ybiw In Complex With 1,5-Anhydroglucitol-6-Phosphate In Escherichia Coli
Organism: Escherichia coli str. k-12 substr. mg1655
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2024-10-02 Classification: LYASE Ligands: 0WK |
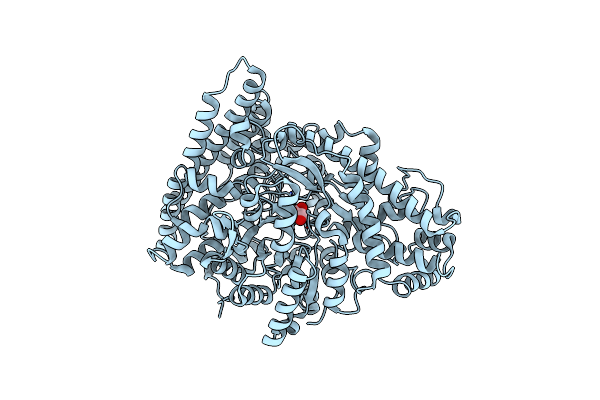 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2024-10-02 Classification: LYASE Ligands: GOL |
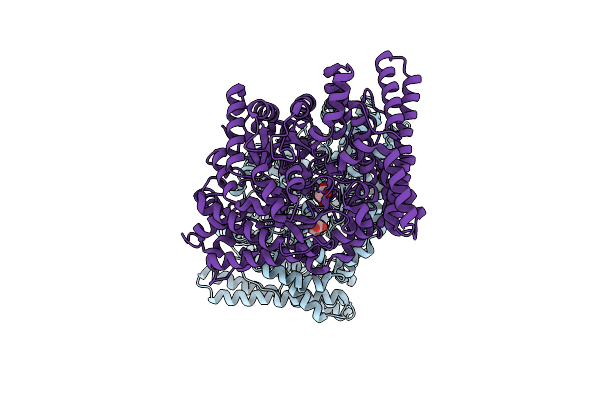 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-10-02 Classification: LYASE Ligands: MLI |
 |
Structure Of The First Otu Domain From Legionella Pneumophila Effector Protein Lota
Organism: Legionella pneumophila
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-12-21 Classification: HYDROLASE Ligands: GOL, IOD |
 |
Structure Of The First Otu Domain From Legionella Pneumophila Effector Protein Lota Bound To K6-Linked Diub
Organism: Homo sapiens, Legionella pneumophila
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2022-12-21 Classification: HYDROLASE |
 |
Organism: Legionella pneumophila subsp. pneumophila str. philadelphia 1
Method: X-RAY DIFFRACTION Resolution:2.74 Å Release Date: 2022-06-15 Classification: HYDROLASE Ligands: SO4 |
 |
Organism: Legionella pneumophila subsp. pneumophila str. philadelphia 1
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2022-06-15 Classification: HYDROLASE Ligands: AR6 |
 |
Substrates Promiscuity Of Xyloglucanases And Endoglucanases Of Glycoside Hydrolase 12 Family
Organism: Aspergillus fischeri
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2020-06-17 Classification: HYDROLASE |
 |
Organism: Aspergillus fischeri
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2020-06-17 Classification: HYDROLASE |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2018-01-31 Classification: TRANSCRIPTION Ligands: CZG, NCO |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2007-05-22 Classification: SIGNALING PROTEIN Ligands: NAG, CA, ZN, ACT |
 |
Crystal Structure Of The Native Form Of Uroporphyrin Iii C-Methyl Transferase From Thermus Thermophilus
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2005-02-15 Classification: TRANSFERASE Ligands: CL, EDO |
 |
Crystal Structure Of Uroporphyrin-Iii C-Methyl Transferase From Thermus Thermophilus Complexed With S-Adenyl Homocysteine
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2005-02-01 Classification: TRANSFERASE Ligands: FLC, SAH |
 |
Complex Between Protein Ser/Thr Phosphatase-1 (Delta) And The Myosin Phosphatase Targeting Subunit 1 (Mypt1)
Organism: Homo sapiens, Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2004-06-22 Classification: HYDROLASE Ligands: MN, PGE |
 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2003-03-04 Classification: TRANSFERASE Ligands: SO4, GMP |
 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2003-02-27 Classification: TRANSFERASE Ligands: SO4, ADN |
 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2003-02-27 Classification: TRANSFERASE Ligands: GOL |
 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2003-02-27 Classification: TRANSFERASE Ligands: SO4, GOL, CL |
 |
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 1998-05-27 Classification: CALCIUM-REGULATED MUSCLE CONTRACTION Ligands: CA |

