Search Count: 16
 |
Crystal Structure Of Chorismate Mutase From Mycobacterium Tuberculosis In Complex With The Cyclic Peptide Inhibitor L2.1 (Triclinic Form)
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2025-02-26 Classification: ISOMERASE Ligands: SO4 |
 |
Crystal Structure Of Chorismate Mutase From Mycobacterium Tuberculosis In Complex With The Cyclic Peptide Inhibitor L2.1 (Monoclinic P Form)
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2025-02-26 Classification: ISOMERASE Ligands: SO4 |
 |
Crystal Structure Of Chorismate Mutase From Mycobacterium Tuberculosis In Complex With The Cyclic Peptide Inhibitor D1.3
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-02-26 Classification: ISOMERASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2021-09-29 Classification: SUGAR BINDING PROTEIN Ligands: GLA, ADP, PO4, MG, NA |
 |
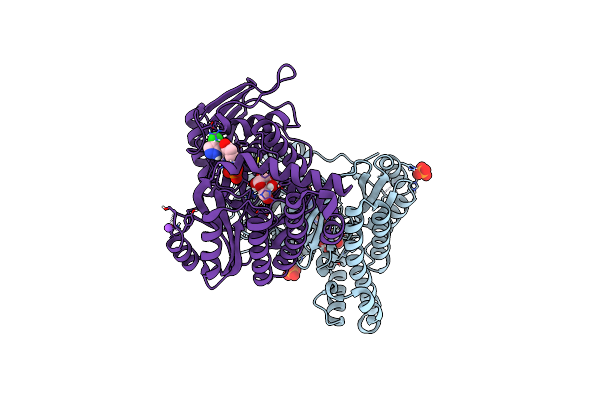
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-09-29 Classification: SUGAR BINDING PROTEIN Ligands: ADP, GLA, PO4, MG, NA, GOL |
 |
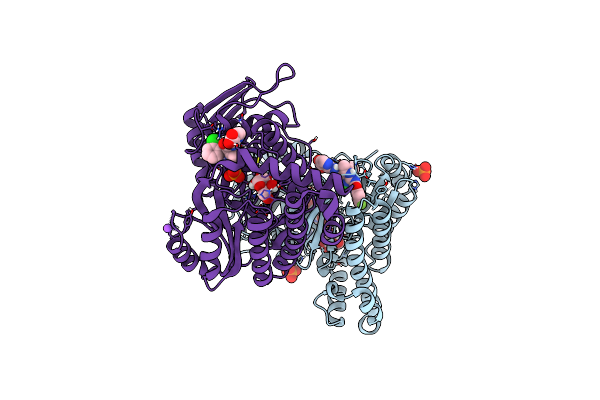
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2021-09-29 Classification: Transferase/Inhibitor Ligands: GLA, 4QI, PO4, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2021-09-29 Classification: Transferase/Inhibitor Ligands: GLA, V3V, NA, PO4 |
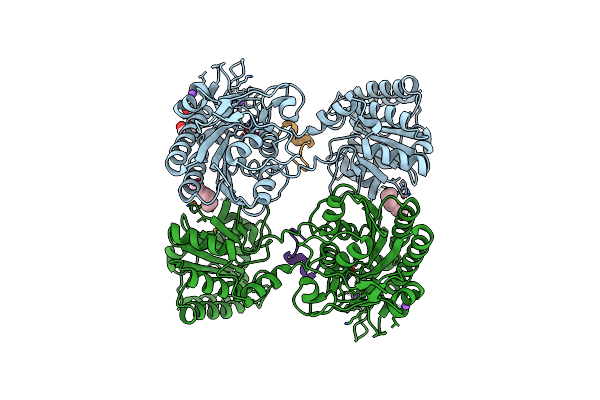 |
1.80A Resolution Structure Of Independent Phosphoglycerate Mutase From C. Elegans In Complex With A Macrocyclic Peptide Inhibitor (Ce-1 Nhoh)
Organism: Caenorhabditis elegans
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-04-07 Classification: ISOMERASE Ligands: ZN, NA, PG4 |
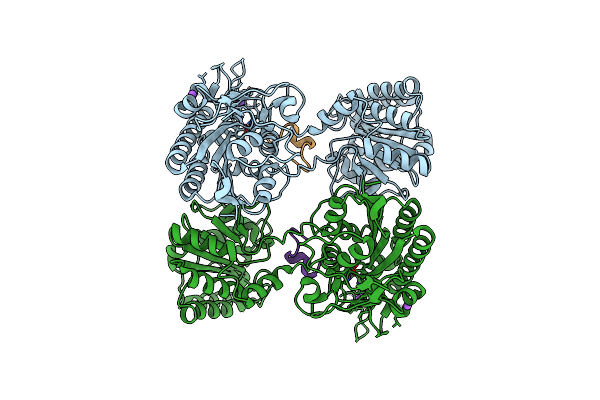 |
2.10A Resolution Structure Of Independent Phosphoglycerate Mutase From C. Elegans In Complex With A Macrocyclic Peptide Inhibitor (Ce-2 Y7F)
Organism: Caenorhabditis elegans
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-04-07 Classification: ISOMERASE Ligands: CL, NA, ZN |
 |
Crystal Structure Of Ldha In Complex With Compound Ncgc00384414-01 At 2.05 A Resolution
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2020-09-23 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE inhibitor Ligands: PO4, P8M, NAI, GOL |
 |
Crystal Structure Of Ldha In Complex With Compound Ncgc00420737-09 At 2.00 A Resolution
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-09-23 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE inhibitor Ligands: NAI, P8V, EDO |
 |
Organism: Anopheles gambiae, Drosophila
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2015-06-17 Classification: HYDROLASE Ligands: ZN, GOL, NHE, CU |
 |
Use Of Synthetic Symmetrization In The Crystallization And Structure Determination Of Cela From Thermotoga Maritima
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2010-12-01 Classification: HYDROLASE Ligands: BR |
 |
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2006-10-17 Classification: HYDROLASE Ligands: SO4 |
 |
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2006-10-17 Classification: HYDROLASE Ligands: SO4, GOL |
 |
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2006-10-17 Classification: HYDROLASE Ligands: TRS |

