Search Count: 12
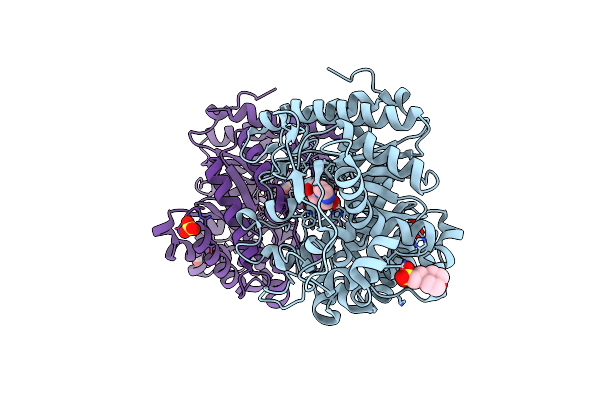 |
Organism: Oryza sativa subsp. japonica
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2020-07-08 Classification: HYDROLASE Ligands: GIM, MES, ZN, SO4 |
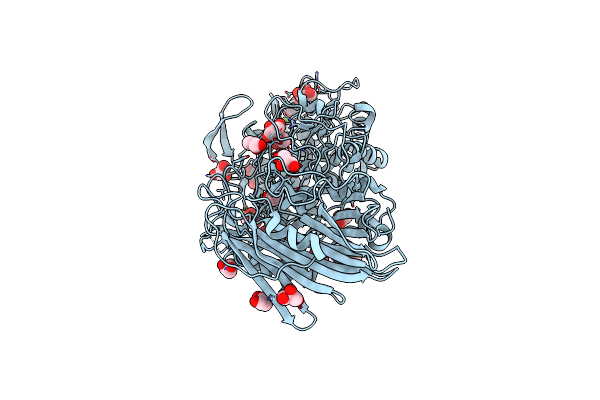 |
Crystal Structure Of Thermoanaerobacterium Xylolyticum Gh116 Beta-Glucosidase
Organism: Thermoanaerobacterium xylanolyticum lx-11
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2016-05-18 Classification: HYDROLASE Ligands: GOL, CA, EDO |
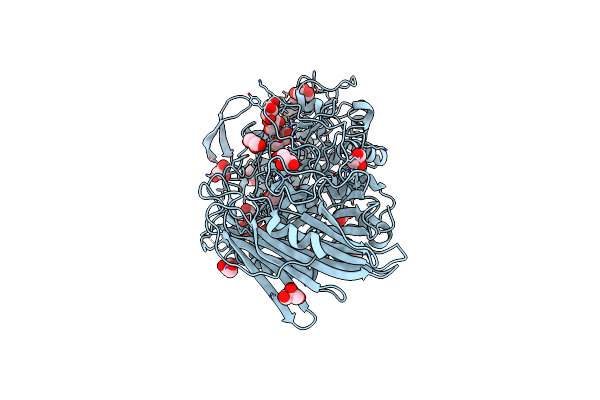 |
Crystal Structure Of Thermoanaerobacterium Xylanolyticum Gh116 Beta-Glucosidase With 2-Deoxy-2-Fluoroglucoside
Organism: Thermoanaerobacterium xylanolyticum lx-11
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2016-05-18 Classification: HYDROLASE Ligands: G2F, GOL, EDO, CA |
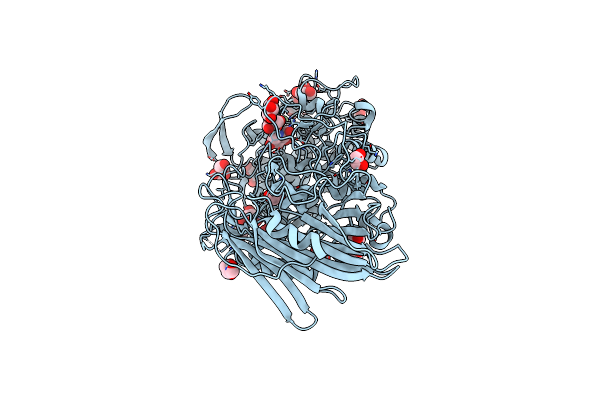 |
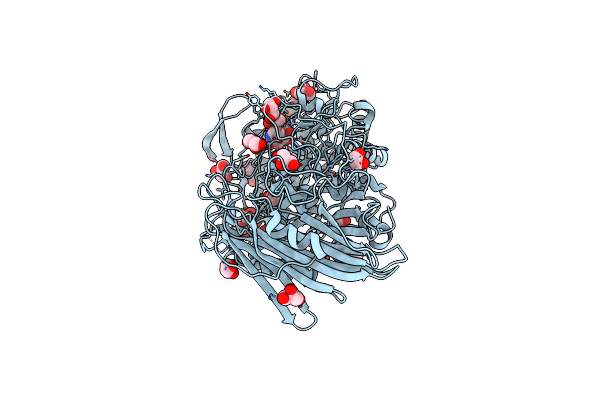
Crystal Structure Of Thermoanaerobacterium Xylanolyticum Gh116 Beta-Glucosidase With Deoxynojirimycin
Organism: Thermoanaerobacterium xylanolyticum lx-11
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2016-05-18 Classification: HYDROLASE Ligands: NOJ, GOL, CA, EDO |
 |
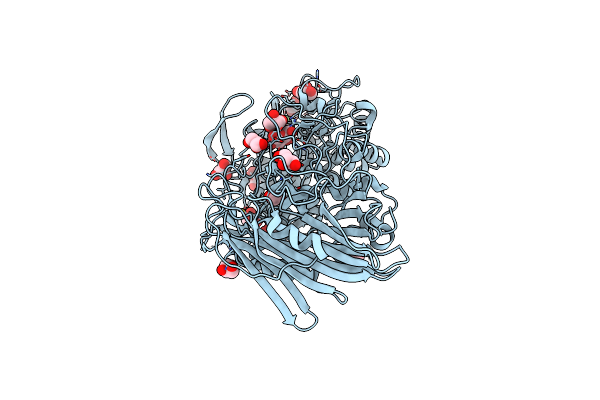
Crystal Structure Of Thermoanaerobacterium Xylanolyticum Gh116 Beta-Glucosidase With Glucoimidazole
Organism: Thermoanaerobacterium xylanolyticum lx-11
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2016-05-18 Classification: HYDROLASE Ligands: GIM, GOL, CA, EDO |
 |
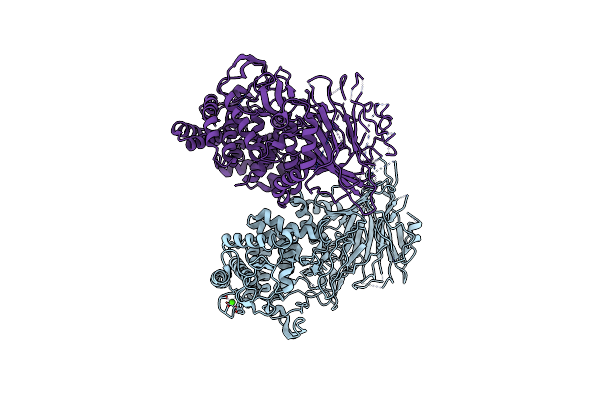
Crystal Structure Of Thermoanaerobacterium Xylanolyticum Gh116 Beta-Glucosidase With Glucose
Organism: Thermoanaerobacterium xylanolyticum lx-11
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2016-05-18 Classification: HYDROLASE Ligands: BGC, GOL, EDO, CA |
 |
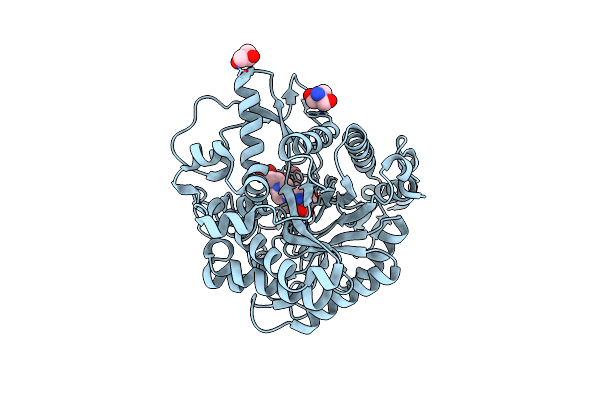
Bacterial Beta-Glucosidase Reveals The Structural And Functional Basis Of Genetic Defects In Human Glucocerebrosidase 2 (Gba2)
Organism: Thermoanaerobacterium xylanolyticum
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2016-05-11 Classification: HYDROLASE Ligands: CA |
 |
Different Transition State Conformations For The Hydrolysis Of Beta-Mannosides And Beta-Glucosides In The Rice Os7Bglu26 Family Gh1 Beta-Mannosidase/Beta-Glucosidase
Organism: Oryza sativa indica group
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-09-16 Classification: HYDROLASE Ligands: MVL, TRS, EPE, GOL |
 |
Different Transition State Conformations For The Hydrolysis Of Beta-Mannosides And Beta-Glucosides In The Rice Os7Bglu26 Family Gh1 Beta-Mannosidase/Beta-Glucosidase
Organism: Oryza sativa indica group
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2015-09-16 Classification: HYDROLASE Ligands: GOL, TRS, EPE, GIM |
 |
Different Transition State Conformations For The Hydrolysis Of Beta-Mannosides And Beta-Glucosides In The Rice Os7Bglu26 Family Gh1 Beta-Mannosidase/Beta-Glucosidase
Organism: Oryza sativa indica group
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2015-09-16 Classification: HYDROLASE Ligands: IFM, GOL, EPE |
 |
Structural Analysis And Insights Into Glycon Specificity Of The Rice Gh1 Os7Bglu26 Beta-D-Mannosidase
Organism: Oryza sativa indica group
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2013-10-09 Classification: HYDROLASE Ligands: EPE, GOL |
 |
Structural Analysis And Insights Into Glycon Specificity Of The Rice Gh1 Os7Bglu26 Beta-D-Mannosidase
Organism: Oryza sativa indica group
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2013-10-09 Classification: HYDROLASE Ligands: GOL, EPE, BMA |

