Search Count: 76
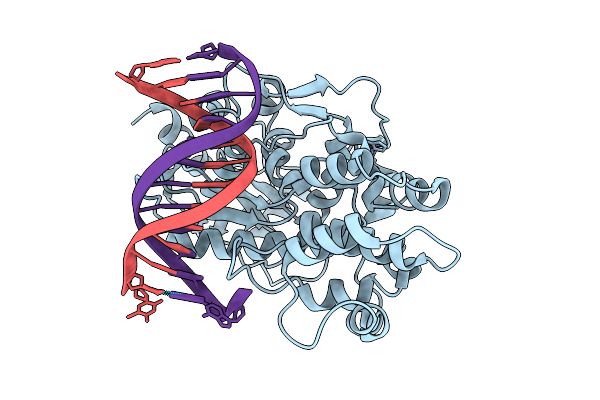 |
Crystal Structure Of Vanc21 In Complex With Its Target Dna (5-Bromouridine Substituted).
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: DNA BINDING PROTEIN/DNA Ligands: ZN |
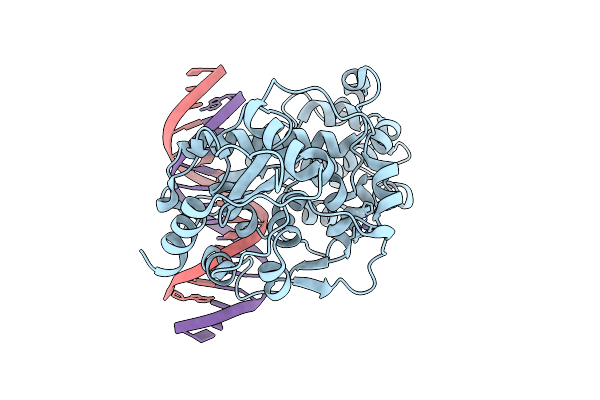 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: DNA BINDING PROTEIN/DNA Ligands: ZN |
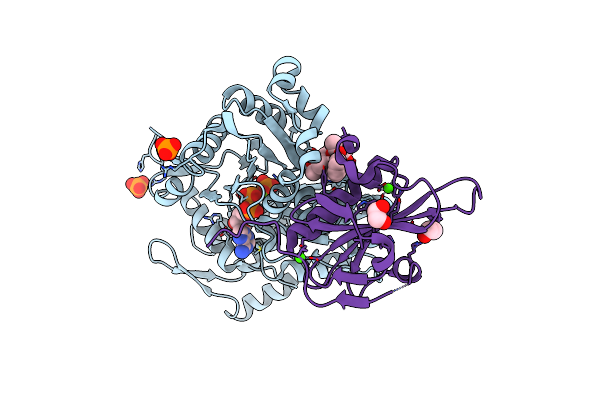 |
Crystal Structure Of Cytochalasin D Bound To A Filamentous Conformation Actin
Organism: Physarum polycephalum, Gallus gallus
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: CYTOSOLIC PROTEIN Ligands: ADP, PO4, MG, CY9, CA, EDO |
 |
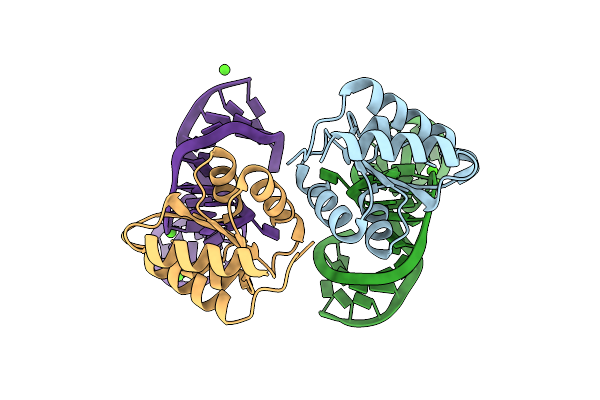
Crystal Structure Of The L7Ae Derivative Protein Ls4 In Complex With Its Co-Evolved Target Cs1 Rna
Organism: Archaeoglobus fulgidus (strain atcc 49558 / dsm 4304 / jcm 9628 / nbrc 100126 / vc-16), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2025-04-02 Classification: RNA BINDING PROTEIN Ligands: MG |
 |
Crystal Structure Of The L7Ae Derivative Protein Ls12 In Complex With Its Co-Evolved Target Cs2 Rna
Organism: Archaeoglobus fulgidus (strain atcc 49558 / dsm 4304 / jcm 9628 / nbrc 100126 / vc-16), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.12 Å Release Date: 2025-04-02 Classification: RNA BINDING PROTEIN Ligands: CA |
 |
Organism: Archaeoglobus fulgidus dsm 4304
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-04-02 Classification: RNA BINDING PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2024-10-23 Classification: CELL CYCLE Ligands: SAH |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.62 Å Release Date: 2024-10-23 Classification: CELL CYCLE Ligands: SAM |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-10-23 Classification: CELL CYCLE Ligands: HOH |
 |
Organism: Arabidopsis thaliana, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: NUCLEAR PROTEIN |
 |
Organism: Arabidopsis thaliana, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: NUCLEAR PROTEIN |
 |
Organism: Arabidopsis thaliana, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: NUCLEAR PROTEIN |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2024-01-31 Classification: STRUCTURAL PROTEIN |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.33 Å Release Date: 2024-01-31 Classification: STRUCTURAL PROTEIN |
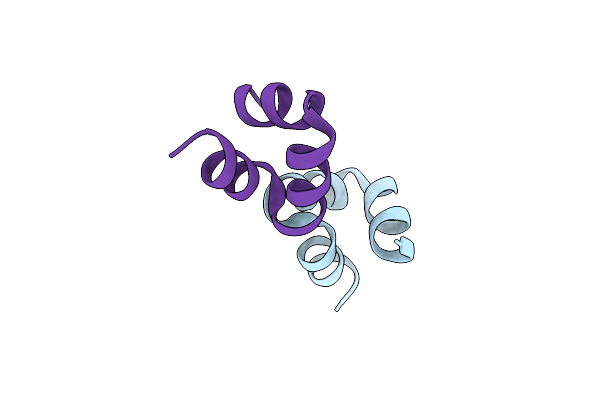 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2024-01-31 Classification: STRUCTURAL PROTEIN |
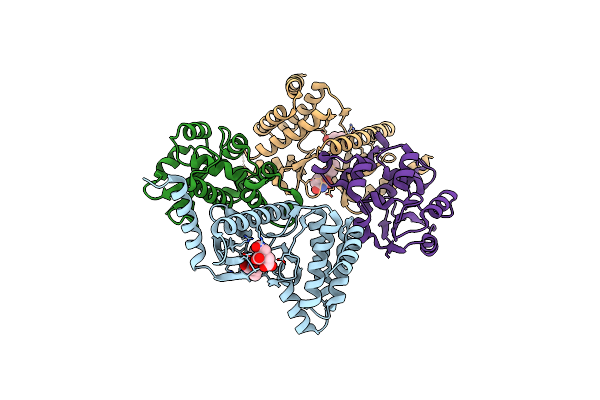 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-01-25 Classification: TRANSFERASE Ligands: DUI, FLC |
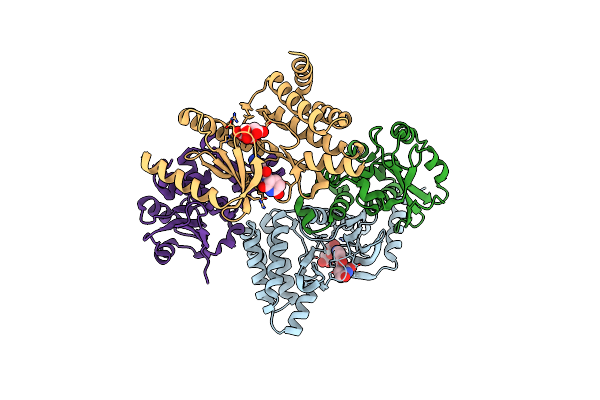 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2023-01-25 Classification: TRANSFERASE Ligands: DQ4, FLC |
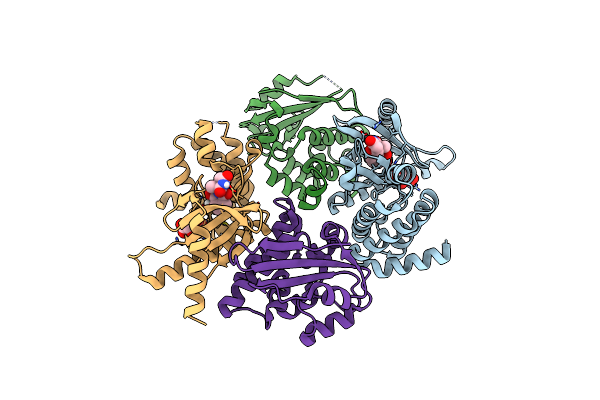 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2022-12-14 Classification: TRANSFERASE Ligands: TAR, DO0 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2022-12-14 Classification: TRANSFERASE Ligands: DVO |
 |
Organism: Homo sapiens, Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.28 Å Release Date: 2022-06-22 Classification: IMMUNE SYSTEM Ligands: EDO, NA, ACT |

