Search Count: 46
 |
Organism: Escherichia coli o127:h6 str. e2348/69
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: DNA BINDING PROTEIN |
 |
Organism: Escherichia coli kly
Method: X-RAY DIFFRACTION Resolution:2.86 Å Release Date: 2024-12-18 Classification: TOXIN |
 |
Crystal Structure Of The E. Coli F-Plasmid Vapbc Toxin-Antitoxin Complex (Vapb T3N, A13P, L16R)
Organism: Escherichia coli kly
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2024-12-18 Classification: TOXIN |
 |
Crystal Structure Of The E. Coli F-Plasmid Vapbc Toxin-Antitoxin Complex (Vapb T3N)
Organism: Escherichia coli kly
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2024-12-18 Classification: TOXIN |
 |
Crystal Structure Of The E. Coli F-Plasmid Vapbc Toxin-Antitoxin Complex (Vapb V5E)
Organism: Escherichia coli kly
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2024-12-18 Classification: TOXIN |
 |
Organism: Shigella sonnei
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2020-08-26 Classification: TOXIN |
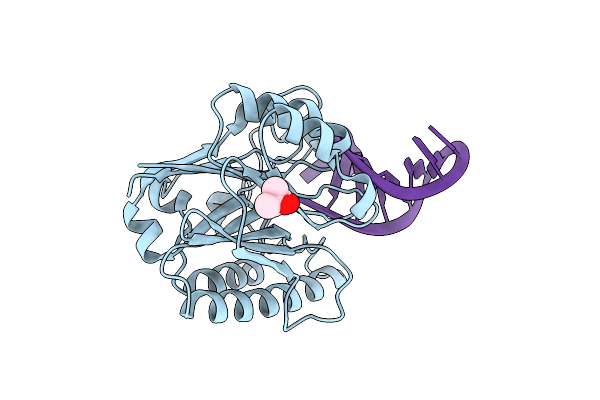 |
Structure Of 3' Phosphatase Nexo (Wt) From Neisseria Bound To Dna Substrate
Organism: Neisseria meningitidis serogroup b (strain mc58), Other sequences
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-10-31 Classification: DNA BINDING PROTEIN Ligands: MPD |
 |
Structure Of 3' Phosphatase Nexo (D146N) From Neisseria Bound To Dna Substrate In Presence Of Magnesium Ion
Organism: Neisseria meningitidis mc58, Other sequences
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2018-10-31 Classification: DNA BINDING PROTEIN Ligands: MG, MPD |
 |
Structure Of 3' Phosphatase Nexo (D146N) From Neisseria Bound To Product Dna Hairpin
Organism: Neisseria meningitidis mc58, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2018-10-31 Classification: DNA BINDING PROTEIN Ligands: MPD, MN |
 |
Structure Of A Fhbp(V1.4):Pora(P1.16) Chimera. Fusion At Fhbp Position 151.
Organism: Neisseria meningitidis, Neisseria meningitidis serogroup c
Method: X-RAY DIFFRACTION Release Date: 2018-02-28 Classification: IMMUNE SYSTEM Ligands: SO4, EDO, CL |
 |
Structure Of A Fhbp(V1.1):Pora(P1.16) Chimera. Fusion At Fhbp Position 294.
Organism: neisseria meningitidis mc58, Neisseria meningitidis serogroup c
Method: X-RAY DIFFRACTION Resolution:3.66 Å Release Date: 2018-02-28 Classification: IMMUNE SYSTEM |
 |
Structure Of A Fhbp(V1.4):Pora(P1.16) Chimera. Fusion At Fhbp Position 309.
Organism: Neisseria meningitidis, Neisseria meningitidis serogroup c
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2018-02-28 Classification: IMMUNE SYSTEM |
 |
Structure Of A Fhbp(V1.1):Pora(P1.16) Chimera. Fusion At Fhbp Position 309.
Organism: neisseria meningitidis mc58, Neisseria meningitidis serogroup c
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2018-02-28 Classification: IMMUNE SYSTEM Ligands: ZN, ACT, EDO |
 |
 |
 |
Nmr Assignment And Structure Of Cssa Thermometer From Neisseria Meningitidis
|
 |
Nmr Assignment And Structure Of Cssa5 (Middle Region) Of Cssa Thermometer From Neisseria Meningitidis
|
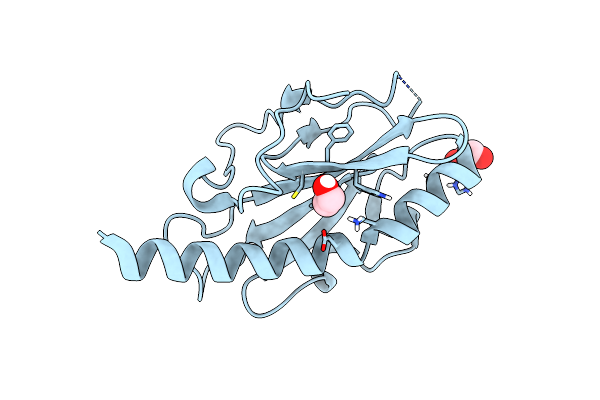 |
Organism: Neisseria meningitidis mc58
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2016-01-13 Classification: CELL ADHESION Ligands: CL, EDO |
 |
Organism: Shigella flexneri
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2015-06-24 Classification: PROTEIN TRANSPORT Ligands: EDO |
 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2014-02-05 Classification: TRANSLATION Ligands: MG, GDP, MES, MPD |

