Search Count: 25
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-17 Classification: MEMBRANE PROTEIN Ligands: A1ETD |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-17 Classification: MEMBRANE PROTEIN Ligands: YN9, CLR, K |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-17 Classification: MEMBRANE PROTEIN Ligands: YN9, CLR, A1ETE, K |
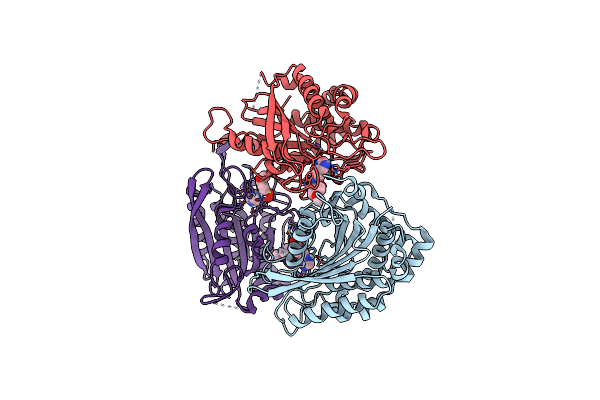 |
Crystal Structure Of Human Glutaminyl Cyclase In Complex With N-(2-(1H-Imidazol-5-Yl)Ethyl)-4-Methoxybenzenesulfonamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: TRANSFERASE Ligands: ZN, A1EFY |
 |
Crystal Structure Of Human Secretory Glutaminyl Cyclase In Complex With The Inhibitor N-(1H-Benzo[D]Imidazol-5-Yl)-1-Phenylmethanesulfonamide (Compound 5)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2025-03-12 Classification: TRANSFERASE Ligands: ZN, A1D93, GOL, DMS, DMF, PEG |
 |
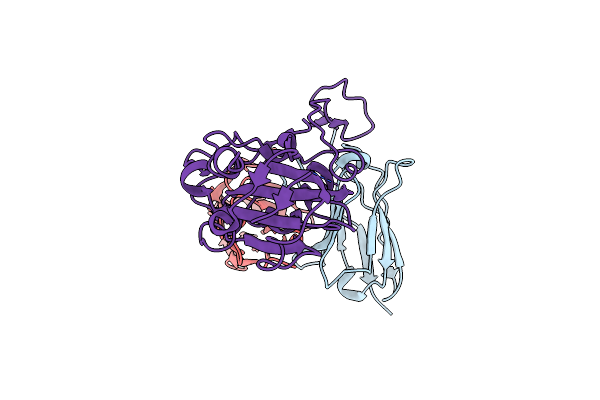
Crystal Structure Of Human Secretory Glutaminyl Cyclase In Complex With The Inhibitor 3-((2-(1H-Imidazol-5-Yl)Ethyl)Carbamoyl)-4-Amino-1,2,5-Oxadiazole 2-Oxide (Compound 13)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.96 Å Release Date: 2025-03-12 Classification: TRANSFERASE Ligands: ZN, A1D94, DMS, GOL, DMF |
 |
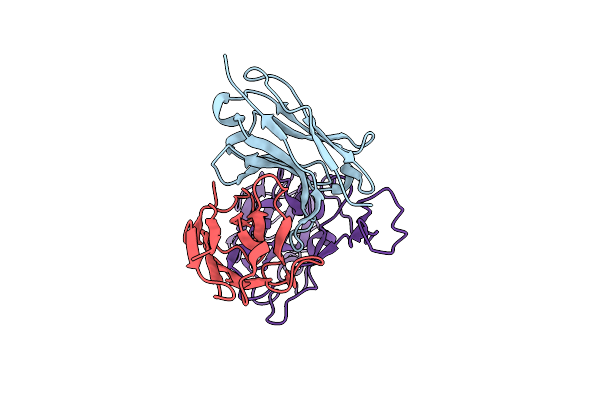
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-09 Classification: VIRAL PROTEIN |
 |
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2024-10-09 Classification: VIRAL PROTEIN |
 |
Pathogen Effectors Which Are Essential To Cause Plant Disease By Manipulating Cellular Processes In The Host
Organism: Phytophthora infestans
Method: X-RAY DIFFRACTION Resolution:3.11 Å Release Date: 2023-07-05 Classification: IMMUNE SYSTEM Ligands: EDO, CD |
 |
Modularity Of Phytophthora Effectors Enables Host Mimicry Of A Principal Phosphatase
Organism: Arabidopsis thaliana, Phytophthora sojae
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2023-07-05 Classification: IMMUNE SYSTEM Ligands: EDO |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-12-28 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Crystal Structure Of Neutralizing Antibody D29 Fab In Complex With Sars-Cov-2 Spike Receptor Binding Domain (Rbd)
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:4.00 Å Release Date: 2022-12-07 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
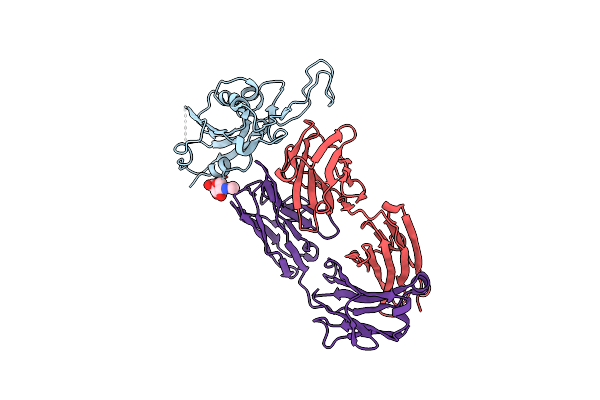 |
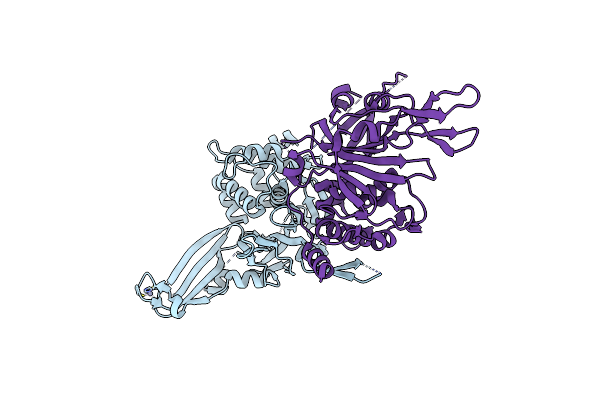
Crystal Structure Of Neutralizing Antibody P1D9 Fab In Complex With Sars-Cov-2 Spike Receptor Binding Domain (Rbd)
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2022-12-07 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
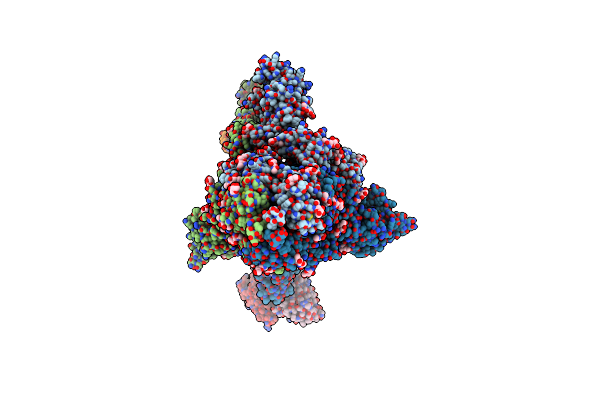 |
Cryo-Em Structure Of Rbd-Directed Neutralizing Antibody P2B4 In Complex With Prefusion Sars-Cov-2 Spike Glycoprotein
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-12-07 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
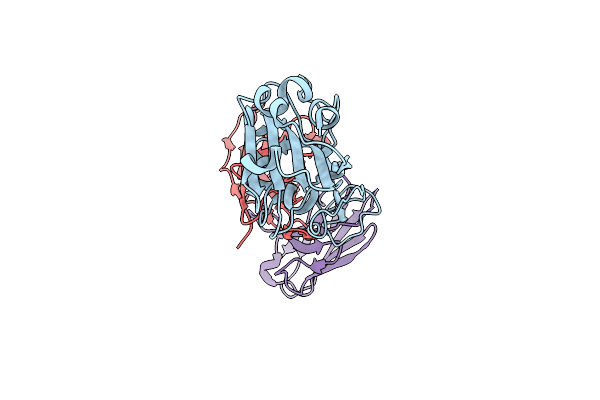 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-06-29 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2022-06-01 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.13 Å Release Date: 2022-06-01 Classification: HYDROLASE Ligands: ZN, AYE |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2020-03-18 Classification: TRANSFERASE/INHIBITOR Ligands: G6P, VMC |
 |
Organism: Pseudoalteromonas sp. cf6-2
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-10-16 Classification: HYDROLASE Ligands: ZN, GOL |
 |
Crystal Structure Of The C-Terminal Guanine Nucleotide Exchange Factor Module Of Human Trio
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2019-02-20 Classification: SIGNALING PROTEIN |

