Search Count: 27
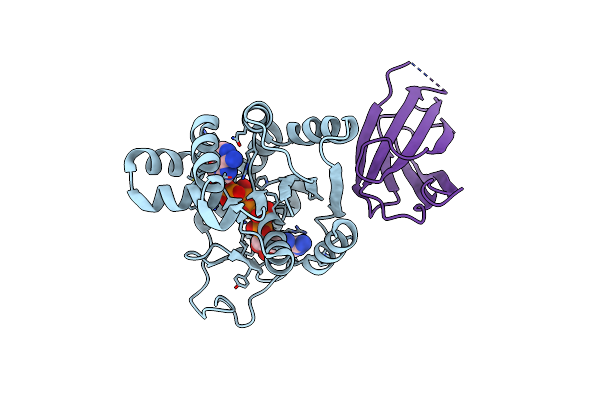 |
Crystal Structure Of The Monobody Cl-1 In Complex With The Escherichia Coli Adenylate Kinase
Organism: Escherichia coli (strain k12), Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-06-04 Classification: PROTEIN BINDING Ligands: AP5, MG |
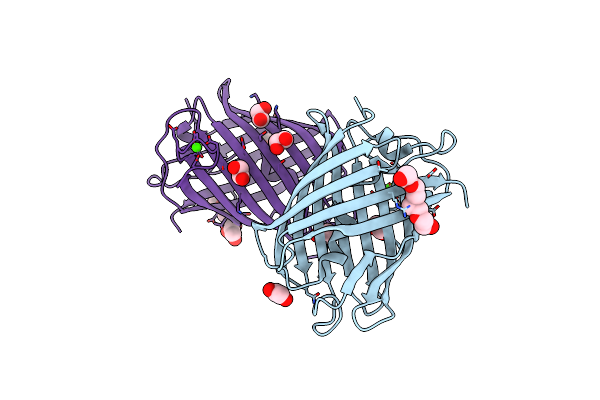 |
Crystal Structure Of Mrfp1 With A Grafted Calcium-Binding Sequence And One Bound Calcium Ion In A Calcium-Containing Solution
Organism: Discosoma, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-06-04 Classification: FLUORESCENT PROTEIN Ligands: CA, EDO, PG4, PGE |
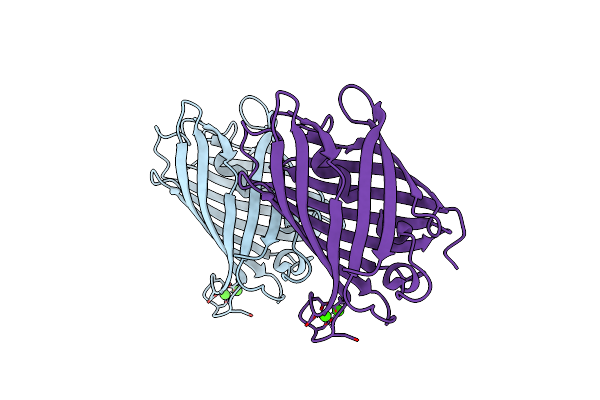 |
Crystal Structure Of Mrfp1 With A Grafted Calcium-Binding Sequence And Two Bound Calcium Ions In A Calcium-Free Solution
Organism: Discosoma, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-06-04 Classification: FLUORESCENT PROTEIN Ligands: CA |
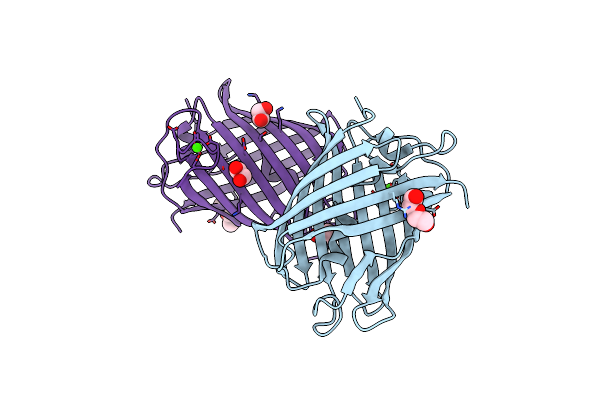 |
Crystal Structure Of Mrfp1 With A Grafted Calcium-Binding Sequence And One Bound Calcium Ion In A Calcium-Free Solution
Organism: Discosoma, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-06-04 Classification: FLUORESCENT PROTEIN Ligands: CA, PEG, EDO |
 |
Crystal Structure Of The Calcium-Free Mrfp1 With A Grafted Calcium-Binding Sequence
Organism: Discosoma, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-06-04 Classification: FLUORESCENT PROTEIN |
 |
Crystal Structure Of The Thermotoga Maritima Cold Shock Protein Mutant Est#13 In Complex With Aacest
Organism: Alicyclobacillus acidocaldarius, Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-01-15 Classification: PROTEIN BINDING Ligands: PMS, PEG, GOL |
 |
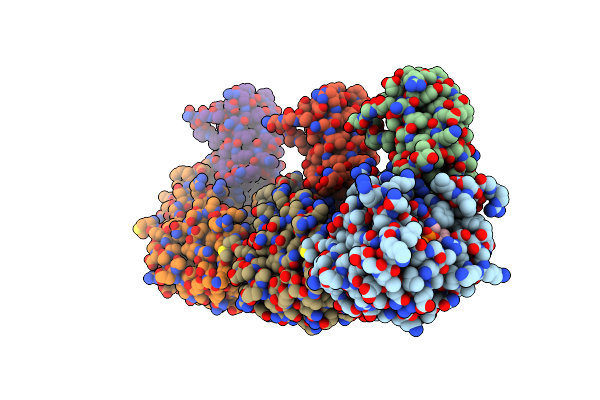
Organism: Klebsiella pneumoniae, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-08-02 Classification: CELL CYCLE Ligands: GDP |
 |
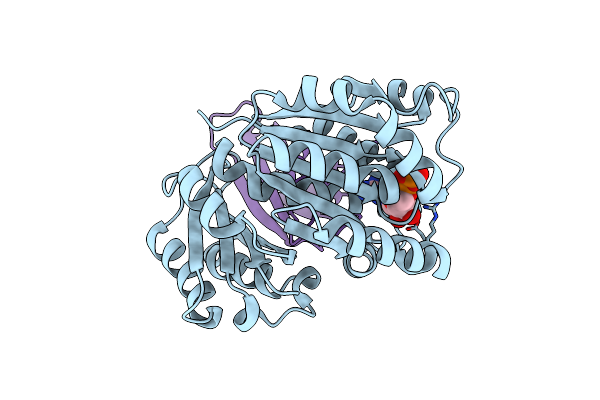
Organism: Klebsiella pneumoniae
Method: ELECTRON MICROSCOPY Release Date: 2023-08-02 Classification: CELL CYCLE Ligands: G2P, K |
 |
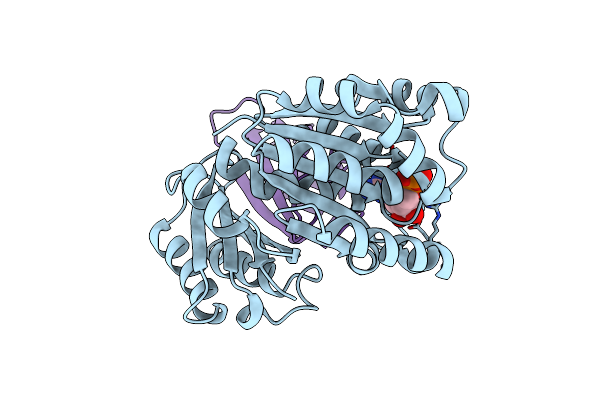
Organism: Klebsiella pneumoniae, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-07-19 Classification: CELL CYCLE Ligands: GDP |
 |
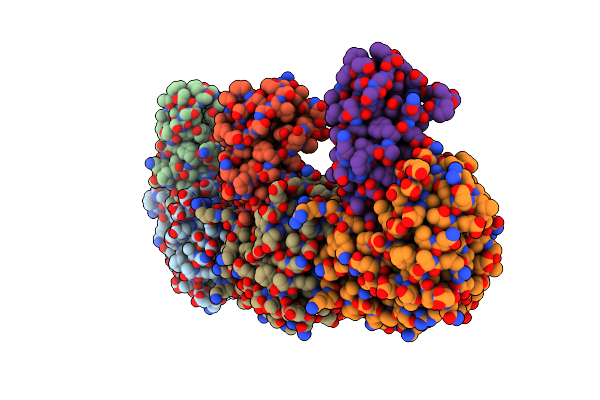
Organism: Klebsiella pneumoniae, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2023-07-19 Classification: CELL CYCLE Ligands: GDP |
 |
Organism: Escherichia coli, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2023-07-19 Classification: CELL CYCLE Ligands: GDP |
 |
Organism: Escherichia coli, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2023-07-19 Classification: CELL CYCLE Ligands: GDP |
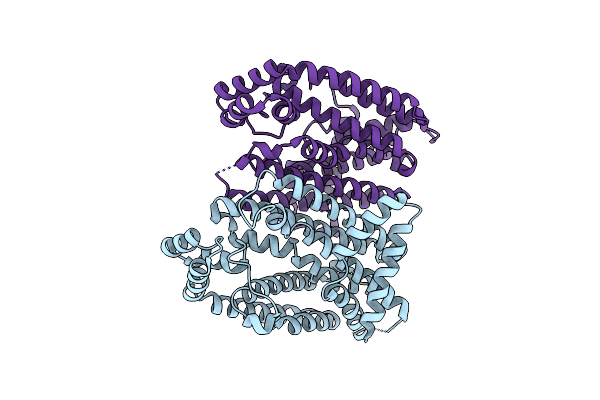 |
Organism: Eucommia ulmoides
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2021-02-17 Classification: PLANT PROTEIN |
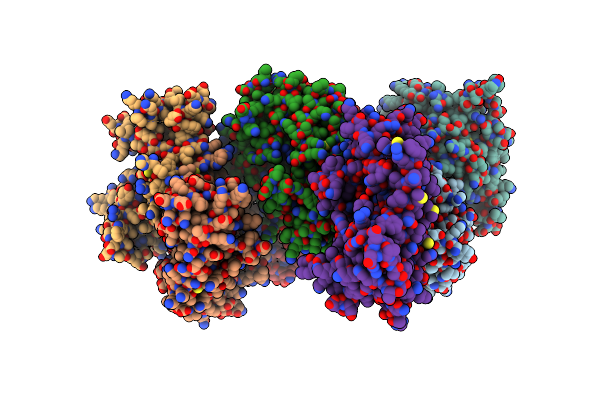 |
Organism: Eucommia ulmoides
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2021-02-17 Classification: PLANT PROTEIN |
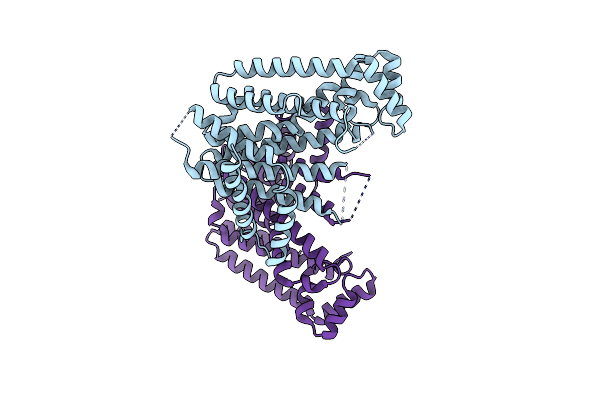 |
Organism: Eucommia ulmoides
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2021-02-17 Classification: PLANT PROTEIN |
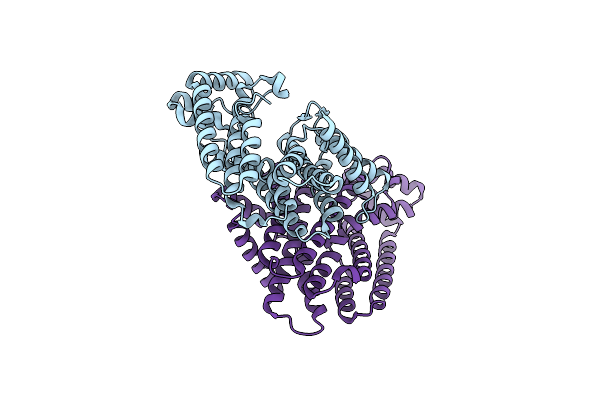 |
Organism: Eucommia ulmoides
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2021-02-17 Classification: PLANT PROTEIN |
 |
Organism: Oryza sativa, Oryza sativa subsp. japonica
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2020-09-16 Classification: PHOTOSYNTHESIS Ligands: SO4, GOL |
 |
Organism: Oryza sativa, Sorghum bicolor
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-09-16 Classification: PHOTOSYNTHESIS Ligands: SO4, GOL |
 |
Organism: Hamamotoa singularis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-09-02 Classification: HYDROLASE Ligands: MAN, NAG, EDO |
 |
Organism: Hamamotoa singularis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-09-02 Classification: HYDROLASE Ligands: NAG, MAN |

