Search Count: 215
 |
Cryo-Em Structure Of Violaxanthin-Chlorophyll-A-Binding Protein With Red Shifted Chl A (Rvcp) From Trachydiscus Minutus At 2.4 Angstrom
Organism: Trachydiscus minutus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: PHOTOSYNTHESIS Ligands: CLA, A1L1F, XAT |
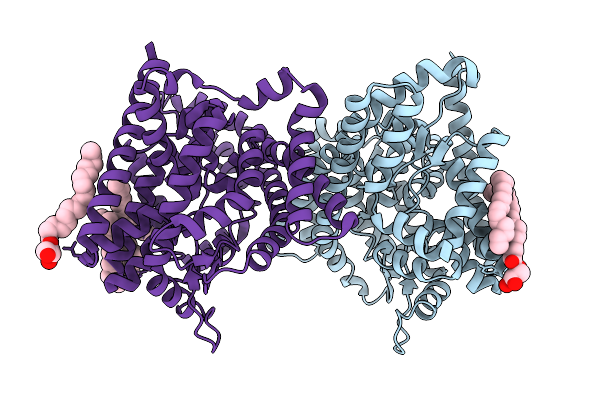 |
Crystal Structure Of Leminorella Grimontii Gatc In The Presence Of D-Xylose
Organism: Leminorella grimontii
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: TRANSPORT PROTEIN Ligands: OLC |
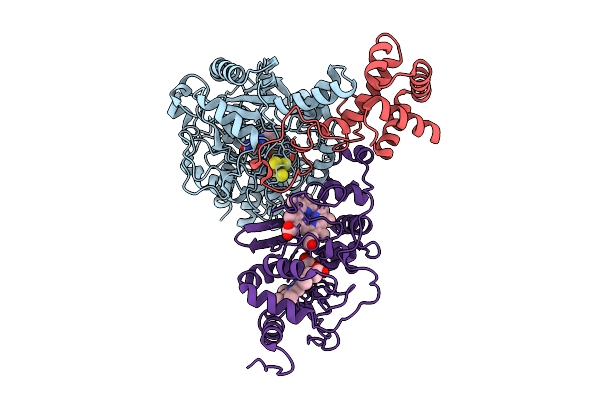 |
Cryo-Em Structure Of Fructose Dehydrogenase Variant From Gluconobacter Japonicus Truncating Heme 1C And C-Terminal Hydrophobic Regions
Organism: Gluconobacter japonicus
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: OXIDOREDUCTASE Ligands: FAD, F3S, HEC |
 |
X-Ray Structure Of Cytochrome P450 Olet From Lacicoccus Alkaliphilus In Complex With Icosanoic Acid
Organism: Lacicoccus alkaliphilus dsm 16010
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: OXIDOREDUCTASE Ligands: HEM, DCR, GOL |
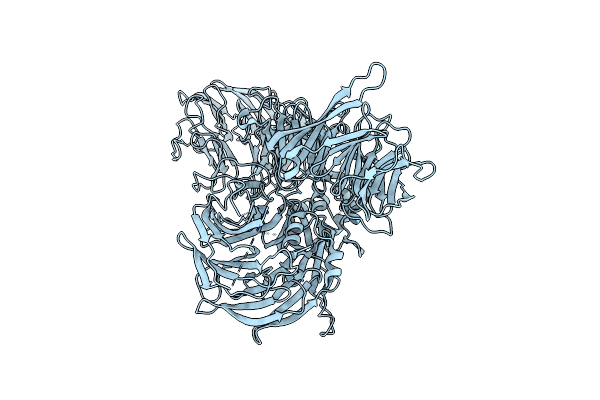 |
Organism: Hepatitis b virus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: VIRAL PROTEIN |
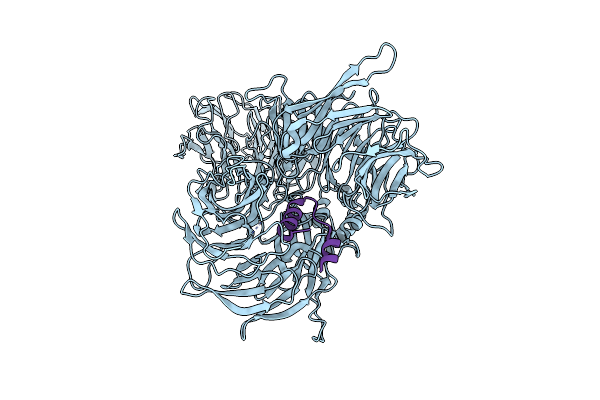 |
Organism: Homo sapiens, Hepatitis b virus
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: VIRAL PROTEIN |
 |
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MOTOR PROTEIN |
 |
Neutron Structure Of Cellulase Cel6A From Phanerochaete Chrysosporium At Room Temperature
Organism: Phanerodontia chrysosporium
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.36 Å, 1.86 Å Release Date: 2025-03-12 Classification: HYDROLASE |
 |
Neutron Structure Of Cellulase Cel6A From Phanerochaete Chrysosporium At Room Temperature, Enzyme-Product Complex
Organism: Phanerodontia chrysosporium
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.40 Å, 1.8 Å Release Date: 2025-03-12 Classification: HYDROLASE Ligands: BGC, NA |
 |
Neutron Structure Of Cellulase Cel6A From Phanerochaete Chrysosporium At Room Temperature, Low-D2O-Solvent
Organism: Phanerodontia chrysosporium
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.40 Å, 2.15 Å Release Date: 2025-03-12 Classification: HYDROLASE |
 |
Neutron Structure Of Cellulase Cel6A From Phanerochaete Chrysosporium At Room Temperature, Enzyme-Product Complex, H2O Solvent
Organism: Phanerodontia chrysosporium
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.80 Å, 2.59 Å Release Date: 2025-03-12 Classification: HYDROLASE Ligands: BGC, NA |
 |
High-Resolution X-Ray Structure Of Cellulase Cel6A From Phanerochaete Chrysosporium At Cryogenic Temperature
Organism: Phanerodontia chrysosporium
Method: X-RAY DIFFRACTION Resolution:0.80 Å Release Date: 2025-03-12 Classification: HYDROLASE Ligands: MPD, ACT |
 |
High-Resolution X-Ray Structure Of Cellulase Cel6A From Phanerochaete Chrysosporium At Cryogenic Temperature, Enzyme-Product Complex
Organism: Phanerodontia chrysosporium
Method: X-RAY DIFFRACTION Resolution:0.85 Å Release Date: 2025-03-12 Classification: HYDROLASE Ligands: BGC, NA, CL, PEG |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-01-29 Classification: GENE REGULATION/DNA |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-01-29 Classification: GENE REGULATION/DNA |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-01-29 Classification: GENE REGULATION/DNA |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-01-29 Classification: GENE REGULATION/DNA |
 |
Neutron Structure Of Ferredoxin-Nadp+ Reductase From Maize Root -Reduced Form
Organism: Zea mays
Method: NEUTRON DIFFRACTION Resolution:1.80 Å Release Date: 2025-01-29 Classification: OXIDOREDUCTASE Ligands: MES, FDA |
 |
Neutron Structure Of Ferredoxin-Nadp+ Reductase From Maize Root -Oxidized Form
Organism: Zea mays
Method: NEUTRON DIFFRACTION Resolution:1.80 Å Release Date: 2025-01-29 Classification: OXIDOREDUCTASE Ligands: FAD, MES |
 |
High Resolution Structure Of Ferredoxin-Nadp+ Reductase From Maize Root - Oxidized Form, Low X-Ray Dose
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2025-01-29 Classification: OXIDOREDUCTASE Ligands: MES, GOL, FAD |

