Search Count: 105
 |
Structure-Based Characterization And Improvement Of An Enzymatic Activity Of Acremonium Alcalophilum Feruloyl Esterase
Organism: Sodiomyces alcalophilus
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2024-04-17 Classification: HYDROLASE Ligands: PEG, MG |
 |
Structure-Based Characterization And Improvement Of An Enzymatic Activity Of Acremonium Alcalophilum Feruloyl Esterase
Organism: Sodiomyces alcalophilus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-04-17 Classification: HYDROLASE Ligands: FER, MG |
 |
Crystal Structure Of Neutralizing Vhh P17 In Complex With Sars-Cov-2 Alpha Variant Spike Receptor-Binding Domain
Organism: Severe acute respiratory syndrome coronavirus 2, Vicugna pacos
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-12-07 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: EDO |
 |
Crystal Structure Of Neutralizing Vhh P17 In Complex With Sars-Cov-2 Alpha Variant Spike Receptor-Binding Domain
Organism: Vicugna pacos
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2022-12-07 Classification: IMMUNE SYSTEM Ligands: CL, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2022-09-21 Classification: LIGASE |
 |
Organism: Vicugna pacos
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-07-20 Classification: IMMUNE SYSTEM Ligands: SO4, EDO |
 |
Cryo-Em Structure Of The Sars-Cov-2 Spike Protein (2-Up Rbd) Bound To Neutralizing Nanobodies P86
Organism: Severe acute respiratory syndrome coronavirus 2, Vicugna pacos
Method: ELECTRON MICROSCOPY Release Date: 2022-07-20 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Ceriporiopsis subvermispora (strain b)
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2022-05-04 Classification: OXIDOREDUCTASE Ligands: NAG, CU, TRS, EDO, XYS, CA |
 |
Hemagglutinin Influenza A Virus (A/Okuda/1957(H2N2) Bound With A Neutralizing Antibody
Organism: Influenza a virus (a/okuda/1957(h2n2)), Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2021-11-24 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Formate Dehydrogenase - Heterodisulfide Reductase - Formylmethanofuran Dehydrogenase Complex From Methanospirillum Hungatei (Hexameric, Composite Structure)
Organism: Methanospirillum hungatei jf-1
Method: ELECTRON MICROSCOPY Release Date: 2021-09-29 Classification: OXIDOREDUCTASE Ligands: SF4, FAD, FES, 9S8, ZN, MO, MGD |
 |
Formate Dehydrogenase - Heterodisulfide Reductase - Formylmethanofuran Dehydrogenase Complex From Methanospirillum Hungatei (Dimeric, Composite Structure)
Organism: Methanospirillum hungatei jf-1
Method: ELECTRON MICROSCOPY Release Date: 2021-09-29 Classification: OXIDOREDUCTASE Ligands: SF4, FAD, FES, 9S8, ZN, MO, MGD |
 |
Formate Dehydrogenase - Heterodisulfide Reductase - Formylmethanofuran Dehydrogenase Complex From Methanospirillum Hungatei (Heterodislfide Reductase Core And Mobile Arm In Conformational State 1, Composite Structure)
Organism: Methanospirillum hungatei jf-1
Method: ELECTRON MICROSCOPY Release Date: 2021-09-29 Classification: OXIDOREDUCTASE Ligands: SF4, FAD, 9S8, FES |
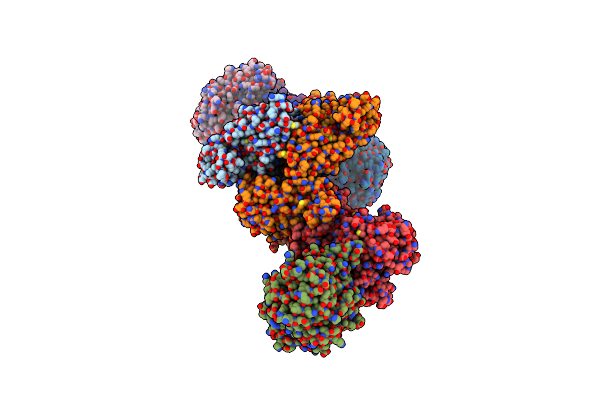 |
Formate Dehydrogenase - Heterodisulfide Reductase - Formylmethanofuran Dehydrogenase Complex From Methanospirillum Hungatei (Heterodisulfide Reductase Core And Mobile Arm In Conformational State 2, Composite Structure)
Organism: Methanospirillum hungatei jf-1
Method: ELECTRON MICROSCOPY Release Date: 2021-09-29 Classification: OXIDOREDUCTASE Ligands: SF4, FAD, 9S8, FES |
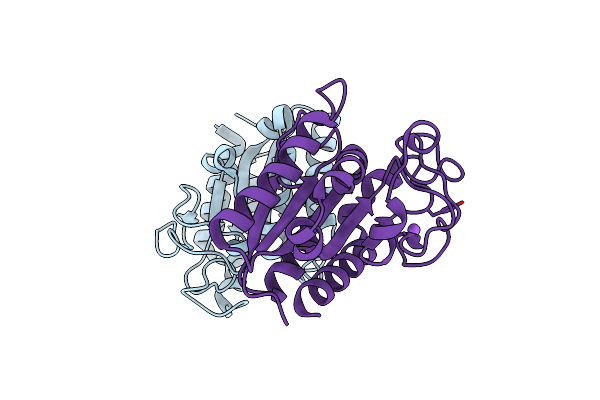 |
A Biodegradable Plastic-Degrading Cutinase-Like Enzyme From The Phyllosphere Yeast Pseudozyma Antarctica
Organism: Pseudozyma antarctica
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-07-15 Classification: HYDROLASE |
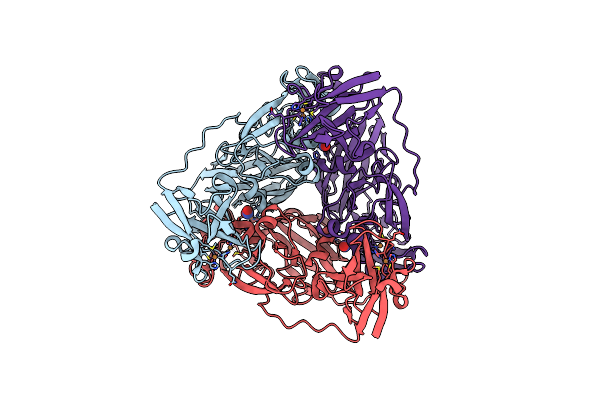 |
Crystal Structure Of Nitrite And No Bound Three-Domain Copper-Containing Nitrite Reductase From Hyphomicrobium Denitrificans Strain 1Nes1
Organism: Hyphomicrobium denitrificans 1nes1
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2020-06-10 Classification: OXIDOREDUCTASE Ligands: CU, NO2, NO |
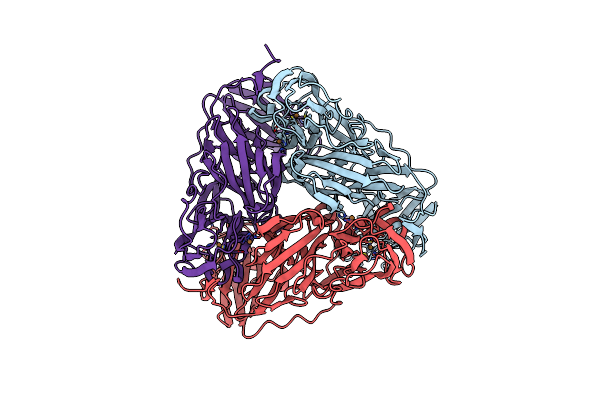 |
Crystal Structure Of As Isolated Three-Domain Copper-Containing Nitrite Reductase From Hyphomicrobium Denitrificans Strain 1Nes1
Organism: Hyphomicrobium denitrificans 1nes1
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2020-06-10 Classification: OXIDOREDUCTASE Ligands: CU |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2020-05-20 Classification: DNA BINDING PROTEIN Ligands: SO4 |
 |
Crystal Structure Of Core Domain Of Four-Domain Heme-Cupredoxin-Cu Nitrite Reductase From Bradyrhizobium Sp. Ors 375
Organism: Bradyrhizobium sp. ors 375
Method: X-RAY DIFFRACTION Resolution:2.87 Å Release Date: 2020-04-22 Classification: OXIDOREDUCTASE Ligands: CU, YB, ER3, TB, YT3, CL, 9JE |
 |
Crystal Structure Of Two-Domain Cu Nitrite Reductase From Bradyrhizobium Sp. Ors 375
Organism: Bradyrhizobium sp. ors 375
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2020-04-22 Classification: OXIDOREDUCTASE Ligands: MES, SO4, CU |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-04-01 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: DU9, ZN, PO4, GOL |

