Search Count: 119
 |
Crystal Structure Of Streptomyces Sp. Sulfotransferase Cpz8 Involved In Caprazamycins Synthesis
Organism: Streptomyces sp.
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: BIOSYNTHETIC PROTEIN Ligands: PO4, MG |
 |
Crystal Structure Of Streptomyces Sp. Sulfotransferase Cpz8 In Complex With Pap, Involved In Caprazamycin Synthesis
Organism: Streptomyces sp.
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: BIOSYNTHETIC PROTEIN Ligands: GOL, A3P, SO4 |
 |
Crystal Structure Of Streptomyces Sp. Sulfotransferase Cpz4 Involved In Caprazamycins Synthesis
Organism: Streptomyces sp.
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: BIOSYNTHETIC PROTEIN Ligands: FMT, MG |
 |
Crystal Structure Of The Reaction Intermediate Of Streptomyces Sp. Sulfotransferase Cpz4 Involved In Caprazamycins Synthesis
Organism: Streptomyces sp.
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: BIOSYNTHETIC PROTEIN Ligands: SO4, MG |
 |
Crystal Structure Of Endo-Processive Xyloglucanase Xeg5A From Aspergillus Oryzae
Organism: Aspergillus oryzae rib40
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: HYDROLASE Ligands: MAN, NAG, GOL |
 |
Crystal Structure Of Endo-Processive Xyloglucanase Xeg5A From Aspergillus Oryzae With Gxg
Organism: Aspergillus oryzae rib40
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: HYDROLASE Ligands: MAN, NAG |
 |
Crystal Structure Of Endo-Processive Xyloglucanase Xeg5A From Aspergillus Oryzae With Xxlg
Organism: Aspergillus oryzae rib40
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: HYDROLASE Ligands: PG4, MAN, NAG, BGC, PEG, MG |
 |
Crystal Structure Of Endo-Processive Xyloglucanase Xeg5A E285A From Aspergillus Oryzae With Gllx/L
Organism: Aspergillus oryzae rib40
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: HYDROLASE Ligands: NAG, MAN |
 |
Organism: Homo sapiens, Bos taurus
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: TRANSFERASE/INHIBITOR Ligands: A1L62 |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2025-04-23 Classification: DE NOVO PROTEIN Ligands: NH4, SO4, ACT, TRS |
 |
Crystal Structure Of L-Ribulose 3-Epimerase From Arthrobacter Globiformis M30
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-02-12 Classification: ISOMERASE Ligands: MG, PEG |
 |
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2025-02-12 Classification: ISOMERASE Ligands: PSJ, FUD, MG, NA, PEG |
 |
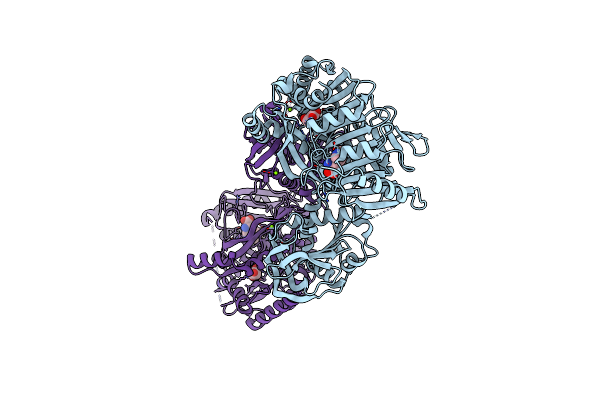
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-01-15 Classification: ELECTRON TRANSPORT, Translocase Ligands: MG, DGT, NDP, ZN, EHZ, HEM, HEC, FMN, SF4, FES |
 |
Structure Of An Acyltransferase Involved In Mannosylerythritol Lipid Formation From Pseudozyma Tsukubaensis In Type A Crystal
Organism: Pseudozyma tsukubaensis
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2024-04-17 Classification: TRANSFERASE Ligands: 1PE |
 |
Structure Of An Acyltransferase Involved In Mannosylerythritol Lipid Formation From Pseudozyma Tsukubaensis In Type B Crystal
Organism: Pseudozyma tsukubaensis
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2024-04-17 Classification: TRANSFERASE Ligands: CL, PGE |
 |
Crystal Structure Of 4-Amino-4-Deoxychorismate Synthase From Streptomyces Venezuelae
Organism: Streptomyces venezuelae
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2023-10-18 Classification: TRANSFERASE Ligands: TRP, MLT, MG, SIN |
 |
Crystal Structure Of 4-Amino-4-Deoxychorismate Synthase From Streptomyces Venezuelae Co-Crystallized With L-Glutamine
Organism: Streptomyces venezuelae
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2023-10-18 Classification: TRANSFERASE Ligands: TRP, SIN, MG, CL |
 |
Crystal Structure Of 4-Amino-4-Deoxychorismate Synthase From Streptomyces Venezuelae Co-Crystallized With Chorismate
Organism: Streptomyces venezuelae
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2023-10-18 Classification: TRANSFERASE Ligands: TRP, SIN, MG |
 |
Crystal Structure Of 4-Amino-4-Deoxychorismate Synthase From Streptomyces Venezuelae With Chorismate
Organism: Streptomyces venezuelae
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2023-10-18 Classification: TRANSFERASE Ligands: TRP, ISJ, GOL, MG, FMT |
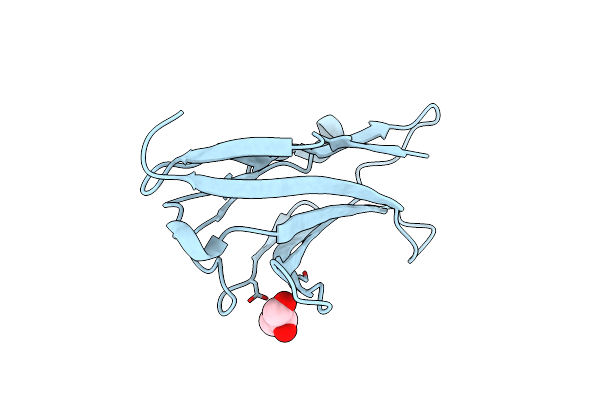 |
Crystal Structure Of The Oligomeric State Of The Extracellular Domain Of Human Myelin Protein Zero(Mpz/P0)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2023-08-30 Classification: CELL ADHESION Ligands: GOL |

