Search Count: 10
 |
The Structure Of Ente With 3-(Prop-2-Yn-1-Yloxy)Benzoic Acid Sulfamoyl Adenosine
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2024-07-31 Classification: LIGASE Ligands: VPT |
 |
The Structure Of Ente With2-Methyl-3-Chloro-Benzoic Acid Sulfamoyl Adenosine
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2024-07-31 Classification: LIGASE Ligands: VQ5 |
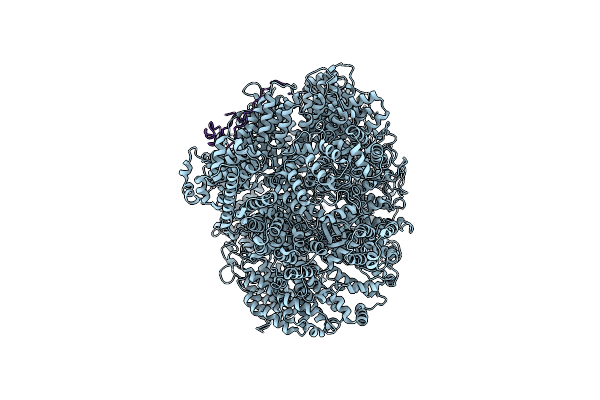 |
Cryo-Em Structure Of The Basal State Of The Artemis:Dna-Pkcs Complex (See Compnd 13/14)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.33 Å Release Date: 2022-07-20 Classification: DNA BINDING PROTEIN |
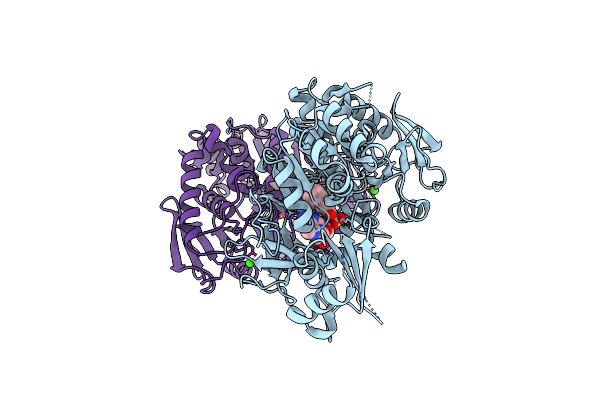 |
Crystal Structure Of Hitb In Complex With (S)-Beta-Phenylalanine Sulfamoyladenosine
Organism: Embleya scabrispora
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2021-03-03 Classification: LIGASE Ligands: 8PZ, CA |
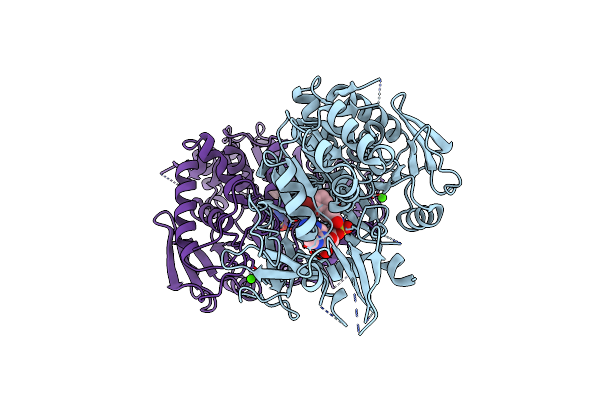 |
Crystal Structure Of Hitb In Complex With (S)-Beta-3-Br-Phenylalanine Sulfamoyladenosine
Organism: Embleya scabrispora
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2021-03-03 Classification: LIGASE Ligands: HFR, CA |
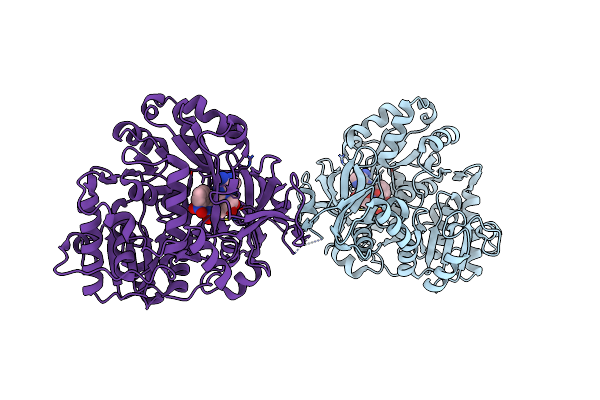 |
Organism: Escherichia coli 1303
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2019-04-17 Classification: LIGASE Ligands: B1U |
 |
Organism: Escherichia coli 1303
Method: X-RAY DIFFRACTION Resolution:2.56 Å Release Date: 2019-04-17 Classification: LIGASE Ligands: B1X |
 |
Organism: Sorangium cellulosum
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2018-06-13 Classification: TRANSFERASE Ligands: SO4, 9EF |
 |
Organism: Homo sapiens, Rattus norvegicus, Bungarus multicinctus
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2017-05-03 Classification: TRANSPORT PROTEIN/TOXIN |
 |
Organism: Mus musculus, Bungarus multicinctus, Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2017-05-03 Classification: TRANSPORT PROTEIN/TOXIN |

