Search Count: 5
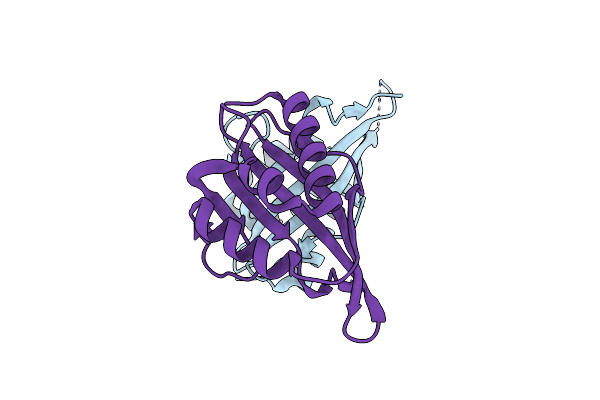 |
Crystal Structure Of A Computationally Designed Retro-Aldolase, Ra110.4 (Cys Free)
Organism: Artificial gene
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2015-04-08 Classification: LYASE |
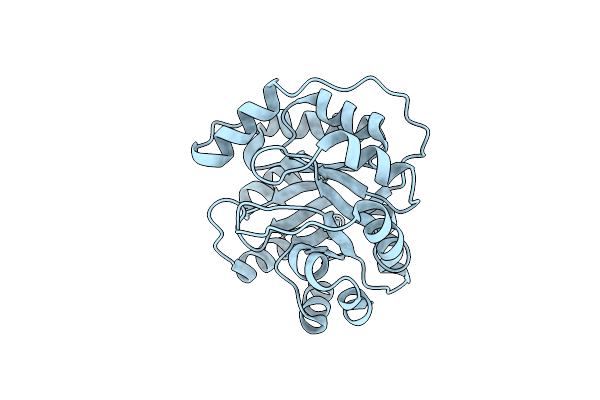 |
Organism: Artificial gene
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2015-04-08 Classification: LYASE |
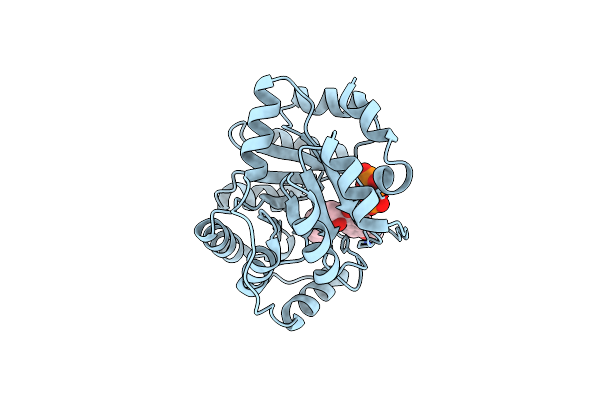 |
Crystal Structure Of A Computationally Designed Retro-Aldolase Covalently Bound To Folding Probe 1 [(6-Methoxynaphthalen-2-Yl)(Oxiran-2-Yl)Methanol]
Organism: Sulfolobus solfataricus
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2014-03-05 Classification: LYASE Ligands: 2V7, BEZ, PO4 |
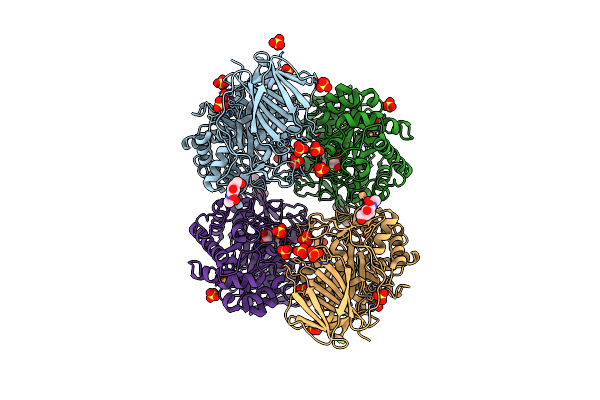 |
The Acid Beta-Glucosidase Active Site Exhibits Plasticity In Binding 3,4,5,6-Tetrahydroxyazepane-Based Inhibitors: Implications For Pharmacological Chaperone Design For Gaucher Disease
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2012-03-14 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: SO4, NAG, 3RI |
 |
The Acid Beta-Glucosidase Active Site Exhibits Plasticity In Binding 3,4,5,6-Tetrahydroxyazepane-Based Inhibitors: Implications For Pharmacological Chaperone Design For Gaucher Disease
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2012-03-14 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: SO4, NAG, 3RK |

