Search Count: 85
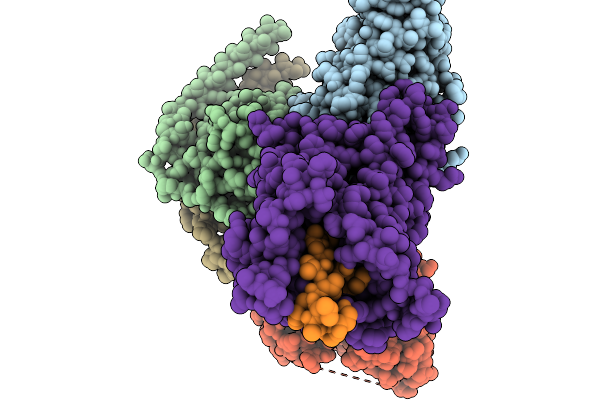 |
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM |
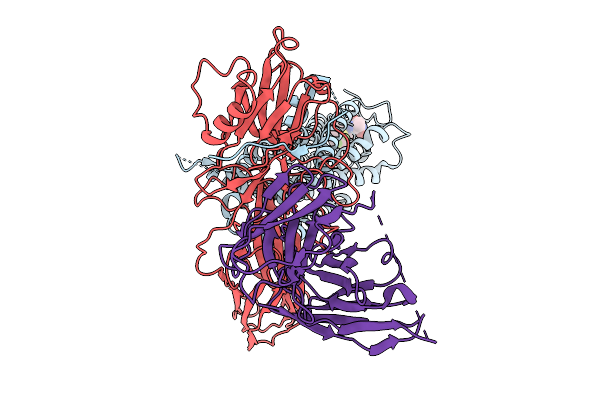 |
Cryo-Em Structure Of The Orphan Gpr52 Beta-Arrestin 1 Complex Bound To Ligand C17
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM Ligands: EN6 |
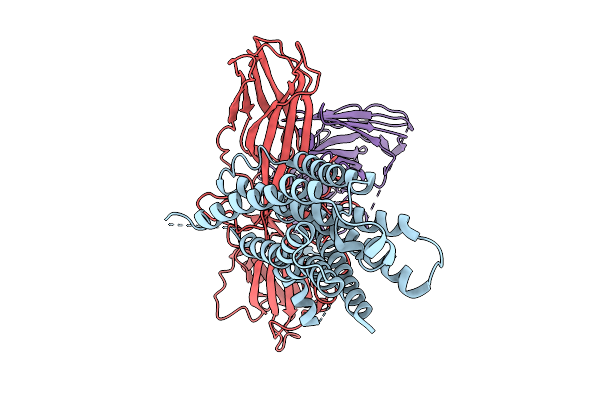 |
Cryo-Em Structure Of The Orphan Gpr52 Bound To Beta-Arrestin 1 In Ligand-Free State
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM |
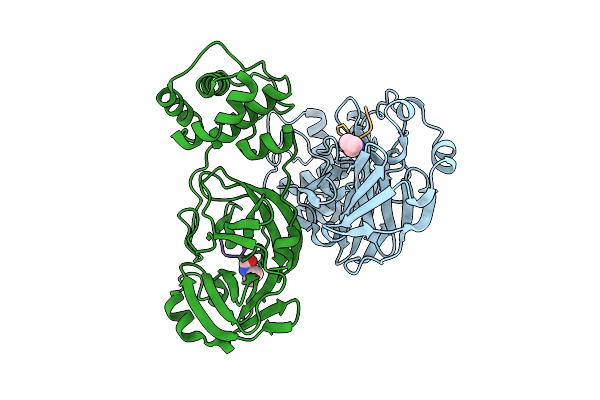 |
Discovery Of An Orally Bioavailable Reversible Covalent Sars-Cov-2 Mpro Inhibitor With Pan-Coronavirus Activity
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: HYDROLASE/INHIBOTOR Ligands: A1L7M |
 |
Discovery Of An Orally Bioavailable Reversible Covalent Sars-Cov-2 Mpro Inhibitor With Pan-Coronavirus Activity
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: HYDROLASE/INHIBITOR |
 |
Discovery Of An Orally Bioavailable Reversible Covalent Sars-Cov-2 Mpro Inhibitor With Pan-Coronavirus Activity
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: HYDROLASE/INHIBITOR |
 |
Discovery Of An Orally Bioavailable Reversible Covalent Sars-Cov-2 Mpro Inhibitor With Pan-Coronavirus Activity
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: HYDROLASE/INHIBITOR Ligands: A1L7M |
 |
Discovery Of An Orally Bioavailable Reversible Covalent Sars-Cov-2 Mpro Inhibitor With Pan-Coronavirus Activity
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: HYDROLASE/INHIBITOR Ligands: A1L7M |
 |
Crystal Structure Of Human Prolyl-Trna Synthetase In Complex With Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: LIGASE Ligands: W2H, EDO |
 |
Organism: Homo sapiens, Streptococcus pneumoniae, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-04-02 Classification: SIGNALING PROTEIN/IMMUNE SYSTEM Ligands: SA0 |
 |
Organism: Homo sapiens, Streptococcus pneumoniae
Method: ELECTRON MICROSCOPY Release Date: 2025-04-02 Classification: SIGNALING PROTEIN Ligands: SA0 |
 |
Organism: Homo sapiens, Streptococcus pneumoniae
Method: ELECTRON MICROSCOPY Release Date: 2025-04-02 Classification: SIGNALING PROTEIN Ligands: SA0 |
 |
Organism: Homo sapiens, Streptococcus pneumoniae
Method: ELECTRON MICROSCOPY Release Date: 2025-04-02 Classification: SIGNALING PROTEIN Ligands: SA0 |
 |
Organism: Thermus antranikianii dsm 12462
Method: ELECTRON MICROSCOPY Release Date: 2025-01-29 Classification: TRANSLOCASE/DNA Ligands: ANP |
 |
Organism: Thermus antranikianii dsm 12462
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: TRANSLOCASE |
 |
Organism: Thermus antranikianii dsm 12462
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: TRANSLOCASE/DNA Ligands: ANP |
 |
Molecular Basis Of The Phosphorothioation-Sensing Antiphage Defence System Dndbcde-Dndi
Organism: Salmonella enterica
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2024-12-11 Classification: DNA BINDING PROTEIN Ligands: PGE, EDO |
 |
Crystal Structure Of Bacterial Prolyl-Trna Synthetase In Complex With Inhibitor Paa-5
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Release Date: 2024-09-11 Classification: LIGASE Ligands: W2H, MG, EDO |
 |
Crystal Structure Of Bacterial Prolyl-Trna Synthetase In Complex With Inhibitor Paa-19
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Release Date: 2024-09-04 Classification: LIGASE Ligands: W1Q, EDO |
 |
Crystal Structure Of Bacterial Prolyl-Trna Synthetase In Complex With Inhibitor Paa-38
Organism: Pseudomonas aeruginosa 148
Method: X-RAY DIFFRACTION Release Date: 2024-09-04 Classification: LIGASE Ligands: W20, SO4, MG, EDO |

