Search Count: 16
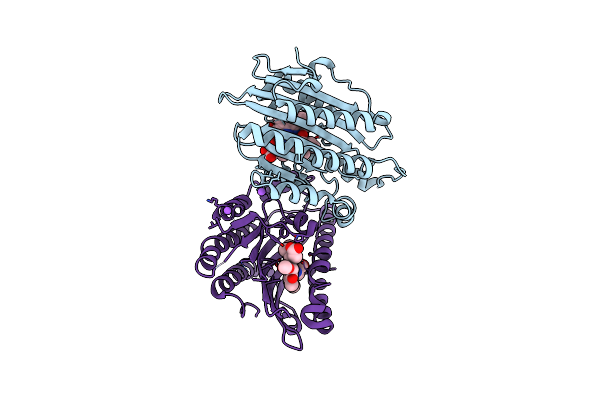 |
Crystal Structure Of Rcc-Bound Red Chlorophyll Catabolite Reductase From Arabidopsis Thaliana
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2010-09-01 Classification: OXIDOREDUCTASE Ligands: RCC, NA |
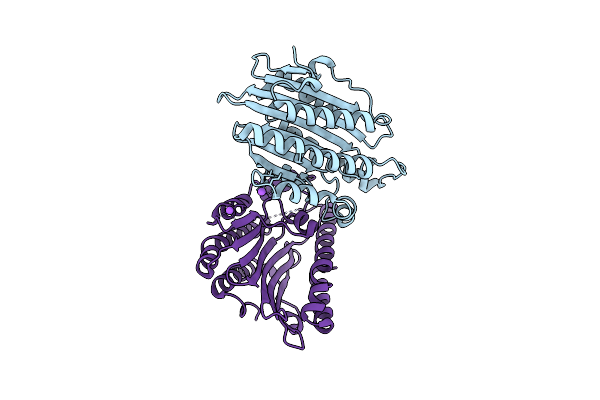 |
F218V Mutant Of The Substrate-Free Form Of Red Chlorophyll Catabolite Reductase From Arabidopsis Thaliana
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2010-09-01 Classification: OXIDOREDUCTASE |
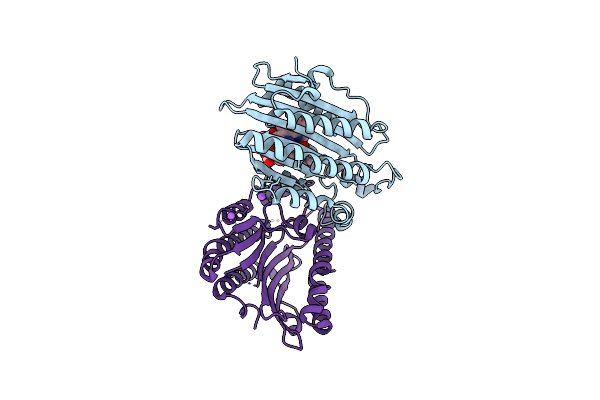 |
F218V Mutant Of The Substrate-Bound Red Chlorophyll Catabolite Reductase From Arabidopsis Thaliana
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2010-09-01 Classification: OXIDOREDUCTASE Ligands: RCC, NA |
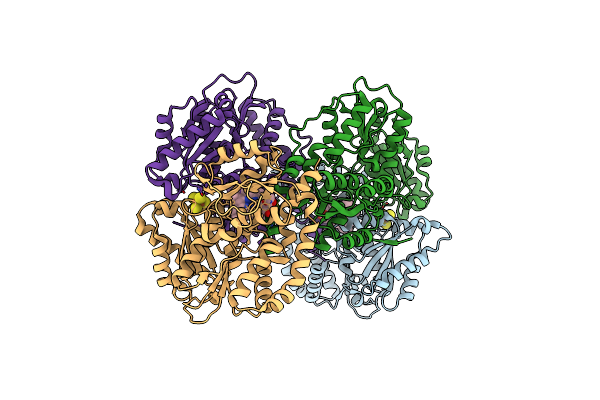 |
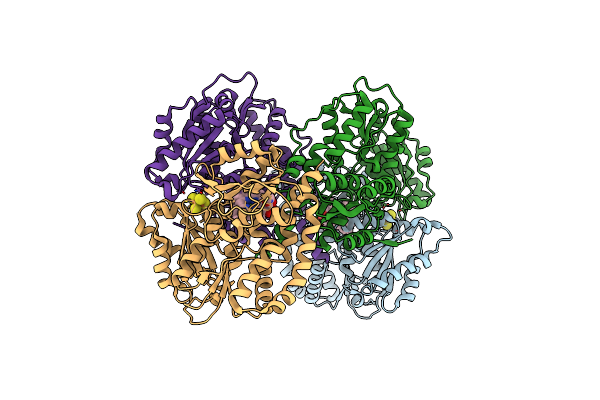
Structure Of The Light-Independent Protochlorophyllide Reductase Catalyzing A Key Reduction For Greening In The Dark
Organism: Rhodobacter capsulatus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2010-04-21 Classification: OXIDOREDUCTASE Ligands: SF4, PMR |
 |
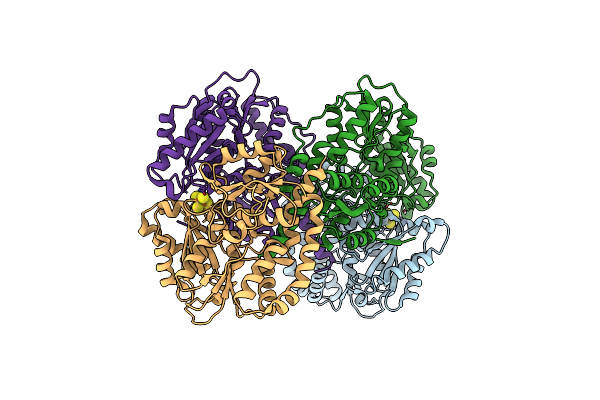
Structure Of The Light-Independent Protochlorophyllide Reductase Catalyzing A Key Reduction For Greening In The Dark
Organism: Rhodobacter capsulatus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2010-04-21 Classification: OXIDOREDUCTASE Ligands: SF4, PMR |
 |
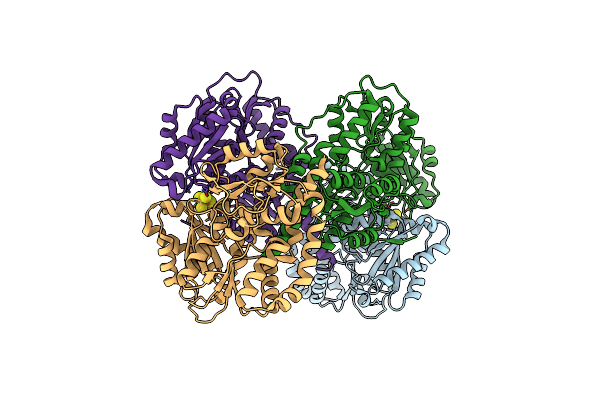
Structure Of The Light-Independent Protochlorophyllide Reductase Catalyzing A Key Reduction For Greening In The Dark
Organism: Rhodobacter capsulatus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2010-04-21 Classification: OXIDOREDUCTASE Ligands: SF4 |
 |
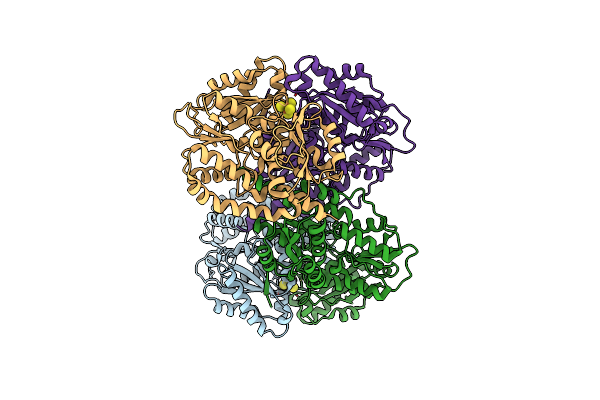
Structure Of The Light-Independent Protochlorophyllide Reductase Catalyzing A Key Reduction For Greening In The Dark
Organism: Rhodobacter capsulatus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2010-04-21 Classification: OXIDOREDUCTASE Ligands: SF4 |
 |
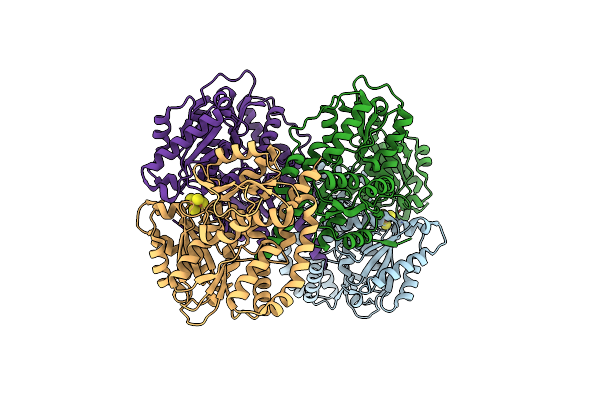
Structure Of The Light-Independent Protochlorophyllide Reductase Catalyzing A Key Reduction For Greening In The Dark
Organism: Rhodobacter capsulatus
Method: X-RAY DIFFRACTION Resolution:2.91 Å Release Date: 2010-04-21 Classification: OXIDOREDUCTASE Ligands: SF4 |
 |
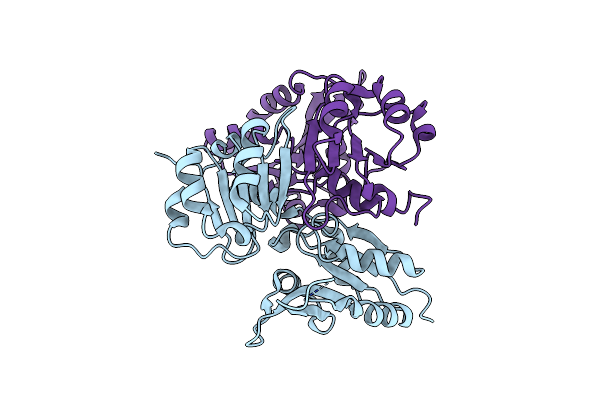
Structure Of The Light-Independent Protochlorophyllide Reductase Catalyzing A Key Reduction For Greening In The Dark
Organism: Rhodobacter capsulatus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2010-04-21 Classification: OXIDOREDUCTASE Ligands: SF4 |
 |
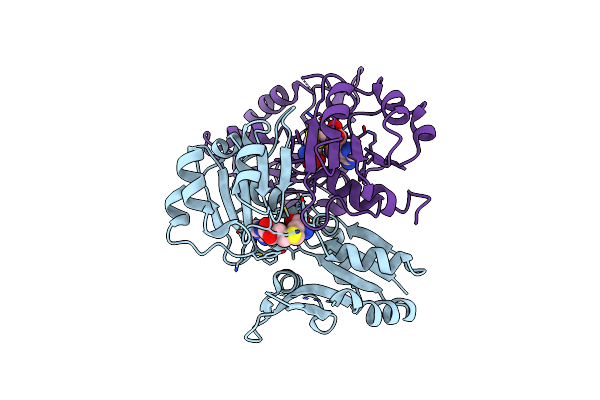
Crystal Structure Of Cbil, A Methyltransferase Involved In Anaerobic Vitamin B12 Biosynthesis
Organism: Chlorobaculum tepidum
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2007-01-16 Classification: TRANSFERASE |
 |
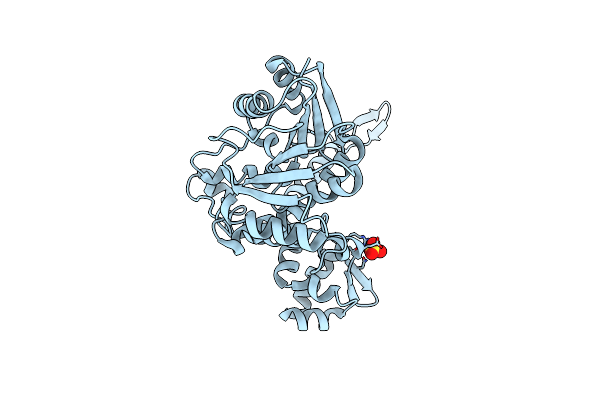
Crystal Structure Of Cbil In Complex With S-Adenosylhomocysteine, A Methyltransferase Involved In Anaerobic Vitamin B12 Biosynthesis
Organism: Chlorobaculum tepidum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2007-01-16 Classification: TRANSFERASE Ligands: SAH |
 |
Organism: Chlorobium tepidum
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2006-07-18 Classification: TRANSFERASE Ligands: SO4 |
 |
Organism: Chlorobium tepidum
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2006-07-18 Classification: TRANSFERASE Ligands: SO4, SAM, GOL |
 |
Organism: Chlorobium tepidum
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2006-07-18 Classification: TRANSFERASE Ligands: SO4, SAH, GOL |
 |
Crystal Structure Of Bchu Complexed With S-Adenosyl-L-Homocysteine And Zn2+
Organism: Chlorobium tepidum
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2006-07-18 Classification: TRANSFERASE Ligands: ZN, SO4, SAH, GOL |
 |
Crystal Structure Of Bchu Complexed With S-Adenosyl-L-Homocysteine And Zn-Bacteriopheophorbide D
Organism: Chlorobium tepidum
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2006-07-18 Classification: TRANSFERASE Ligands: ZN, SO4, SAH, GOL |

