Search Count: 16
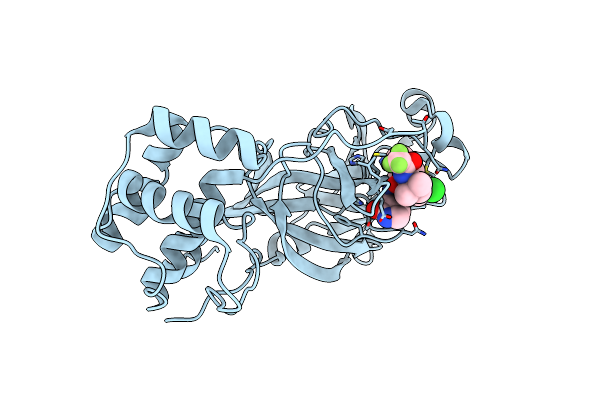 |
Crystal Structure Of Sars-Cov-2 Main Protease In Complex With An Inhibitor Tkb-277-5Cl
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-06-04 Classification: VIRAL PROTEIN Ligands: A1B7C |
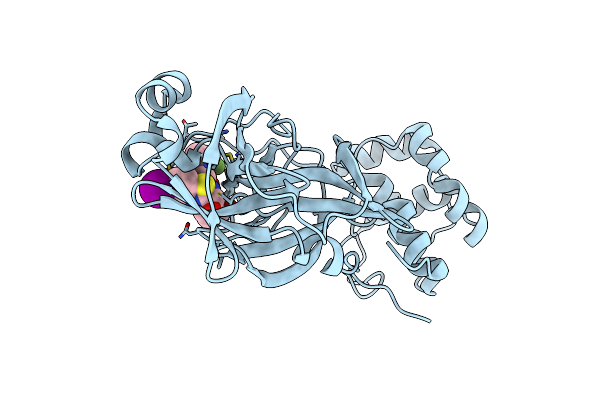 |
Crystal Structure Of Sars-Cov-2 Main Protease In Complex With An Inhibitor Tkb-280-5I
Organism: Severe acute respiratory syndrome coronavirus
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2025-06-04 Classification: VIRAL PROTEIN Ligands: A1B7A |
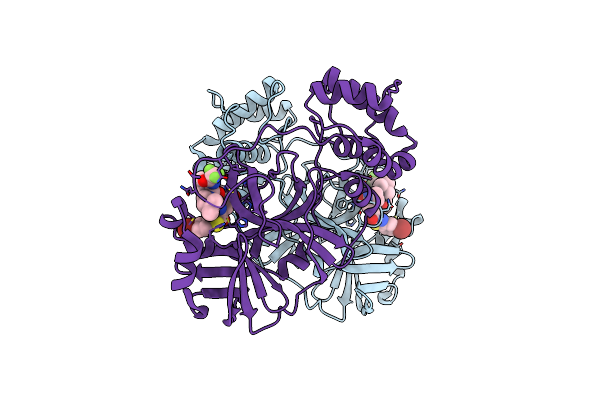 |
Crystal Structure Of Sars-Cov-2 Main Protease In Complex With An Inhibitor Tkb-276-5Br
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2025-06-04 Classification: VIRAL PROTEIN Ligands: A1B7B |
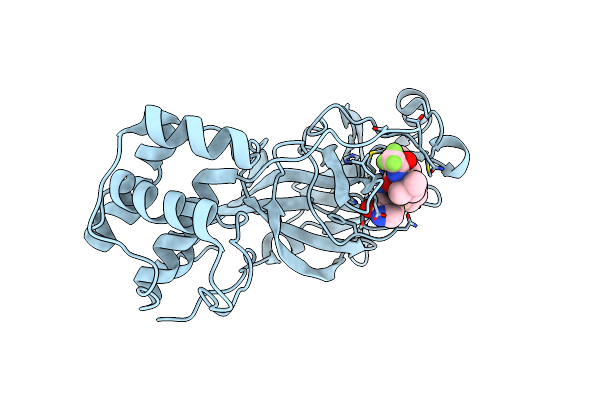 |
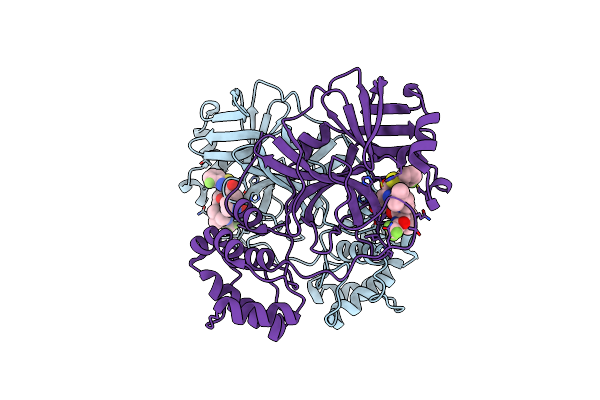
Crystal Structure Of Sars-Cov-2 Main Protease (Authentic Protein) In Complex With An Inhibitor Tkb-245
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-04-24 Classification: VIRAL PROTEIN, HYDROLASE/INHIBITOR Ligands: T2L |
 |
Crystal Structure Of Sars-Cov-2 Main Protease E166V Mutant In Complex With An Inhibitor Tkb-245
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2024-04-24 Classification: VIRAL PROTEIN, HYDROLASE/INHIBITOR Ligands: T2L |
 |
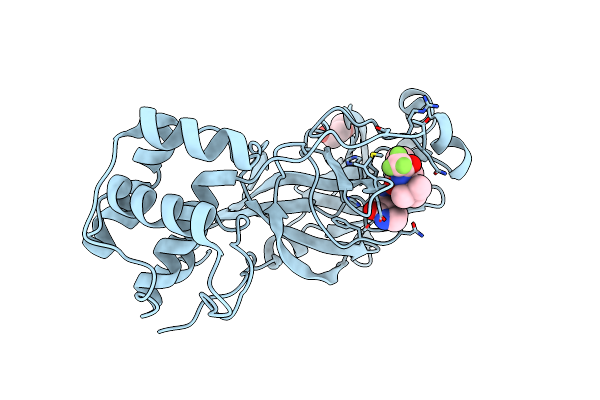
Crystal Structure Of Sars-Cov-2 Main Protease A191T Mutant In Complex With An Inhibitor 5H
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2024-04-24 Classification: VIRAL PROTEIN, HYDROLASE/INHIBITOR Ligands: V7G |
 |
Crystal Structure Of Sars-Cov-2 Main Protease A191T Mutant In Complex With An Inhibitor Nirmatrelvir
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2024-04-24 Classification: VIRAL PROTEIN, HYDROLASE/INHIBITOR Ligands: 4WI, PEG |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-01-24 Classification: VIRAL PROTEIN |
 |
Crystal Structure Of Sars-Cov-2 Main Protease E166V Mutant In Complex With An Inhibitor Tkb-272
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2023-12-20 Classification: VIRAL PROTEIN Ligands: WOK, PEG |
 |
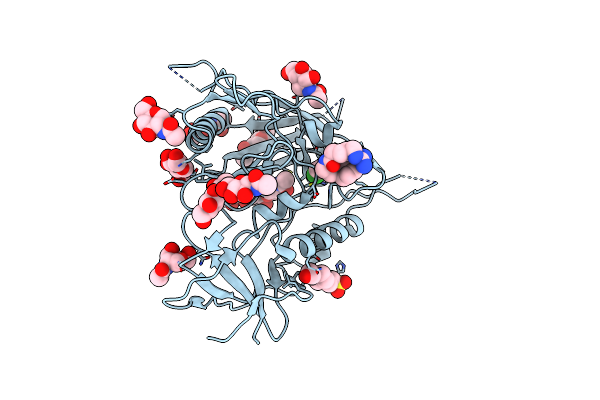
Crystal Structure Of Sars-Cov-2 Main Protease In Complex With An Inhibitor Tkb-272
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2023-12-13 Classification: VIRAL PROTEIN Ligands: WOK, PEG |
 |
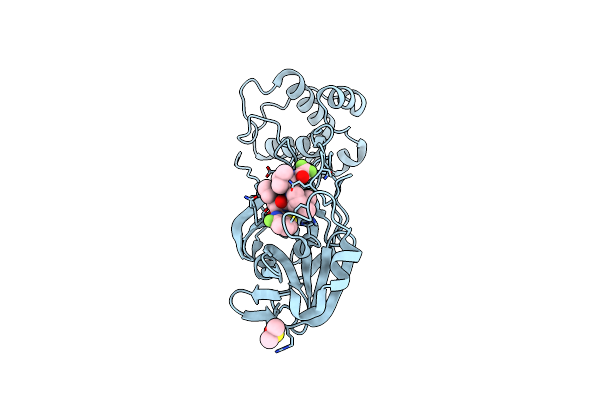
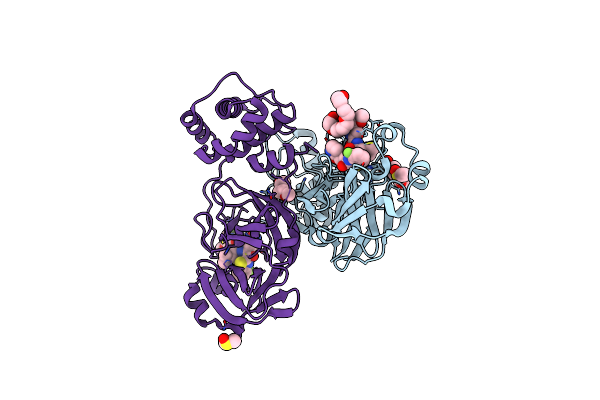
Crystal Structure Of Hiv-1 Lm/Ht Clade A/E Crf01 Gp120 Core In Complex With Yir-821
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2023-01-11 Classification: VIRAL PROTEIN Ligands: NAG, R5K, EPE |
 |
Crystal Structure Of Sars-Cov-2 Main Protease In Complex With An Inhibitor Tkb-245
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2022-09-21 Classification: HYDROLASE/INHIBITOR Ligands: T2L, DMS |
 |
Crystal Structure Of Sars-Cov-2 Main Protease In Complex With An Inhibitor Tkb-198
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2022-08-24 Classification: HYDROLASE/INHIBITOR Ligands: 1PE, MES, DMS, PG0, T1X |
 |
Crystal Structure Of Sars-Cov-2 Main Protease In Complex With Inhibitor Tkb-248
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-08-24 Classification: HYDROLASE/INHIBITOR Ligands: T43, DMS, ZN |
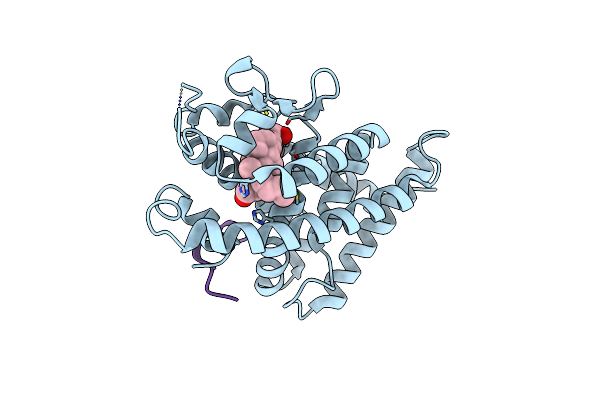 |
A New Class Of Vitamin D Receptor Ligands That Induce Structural Rearrangement Of The Ligand-Binding Pocket
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:3.01 Å Release Date: 2009-02-17 Classification: TRANSCRIPTION Ligands: JB1 |
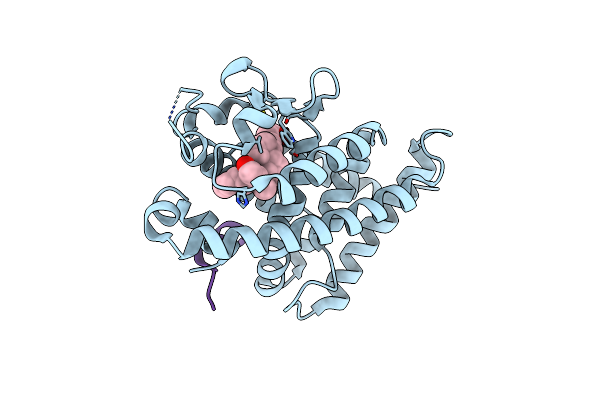 |
A New Class Of Vitamin D Receptor Ligands That Induce Structural Rearrangement Of The Ligand-Binding Pocket
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2009-02-17 Classification: TRANSCRIPTION Ligands: JC1 |

