Search Count: 150
 |
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: OXIDOREDUCTASE Ligands: FDA, NAD |
 |
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: OXIDOREDUCTASE Ligands: FAD, NAI |
 |
Organism: Sus scrofa
Method: NEUTRON DIFFRACTION Release Date: 2025-11-19 Classification: OXIDOREDUCTASE Ligands: FAD, GOL |
 |
Organism: Sus scrofa
Method: NEUTRON DIFFRACTION Release Date: 2025-11-19 Classification: OXIDOREDUCTASE Ligands: FAD, GOL |
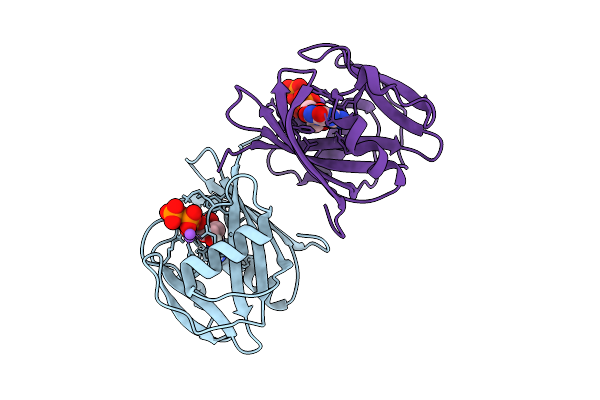 |
Neutron Crystal Structure Of Human Mth1(G2K/C87A/C104S Mutant) In Complex With 8-Oxo-Dgtp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Release Date: 2025-07-30 Classification: HYDROLASE Ligands: 8DG, NA |
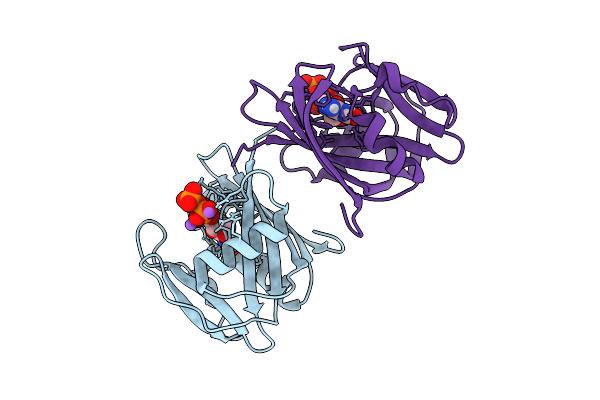 |
Neutron Crystal Structure Of Human Mth1(G2K/C87A/C104S Mutant) In Complex With 2-Oxo-Datp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Release Date: 2025-07-30 Classification: HYDROLASE Ligands: 6U4, NA |
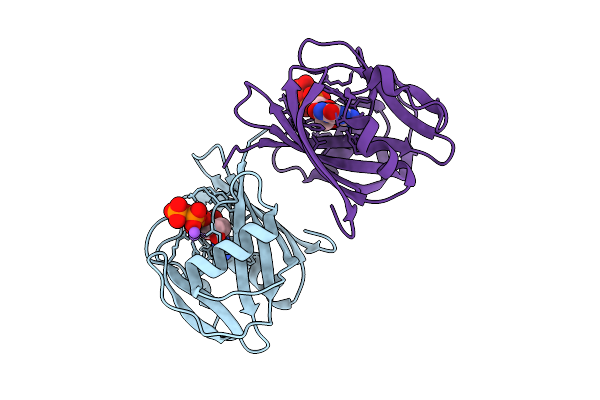 |
8-Oxo-Dgtp Hydrolysis In Human Mth1(G2K Mutant) Crystal Using Mn2+: The Es Complex
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: HYDROLASE Ligands: 8DG, NA |
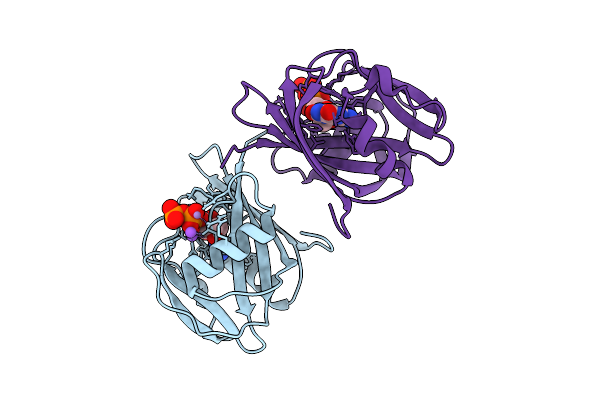 |
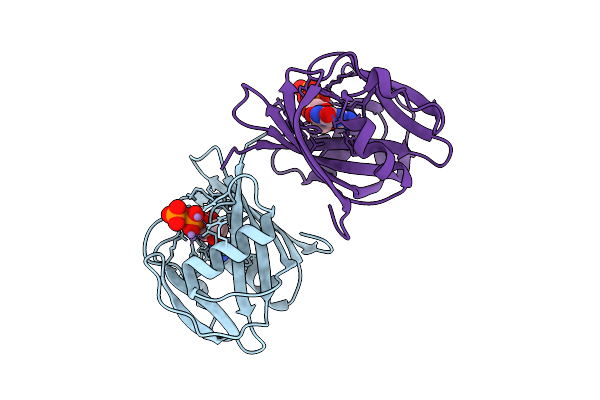
8-Oxo-Dgtp Hydrolysis In Human Mth1(G2K Mutant) Crystal Using Mn2+: The Es-3M Complex
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: HYDROLASE Ligands: 8DG, NA, MN |
 |
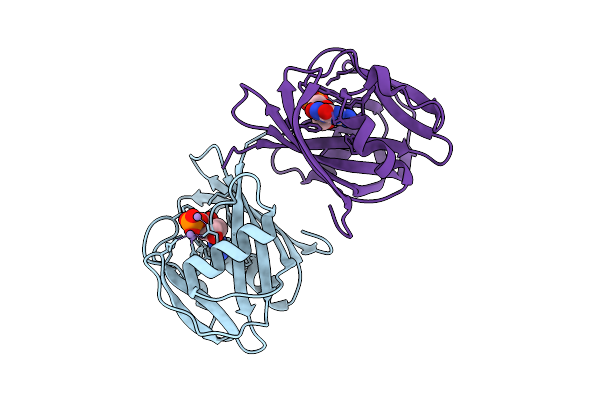
8-Oxo-Dgtp Hydrolysis In Human Mth1(G2K Mutant) Crystal Using Mn2+: The Es/Ep-3M Complex
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: HYDROLASE Ligands: MN, 8DG, 8OG |
 |
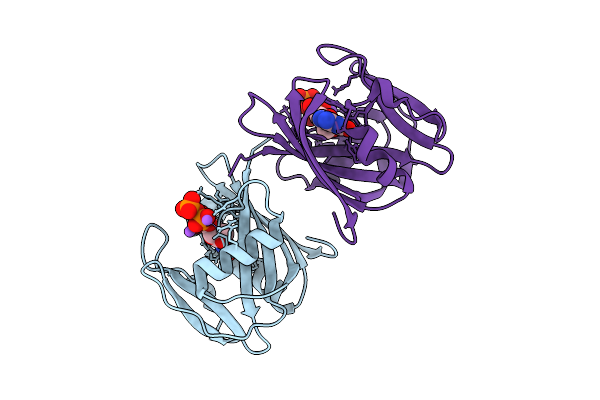
8-Oxo-Dgtp Hydrolysis In Human Mth1(G2K Mutant) Crystal Using Mn2+: The Ep-M Complex
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: HYDROLASE Ligands: 8OG, MN |
 |
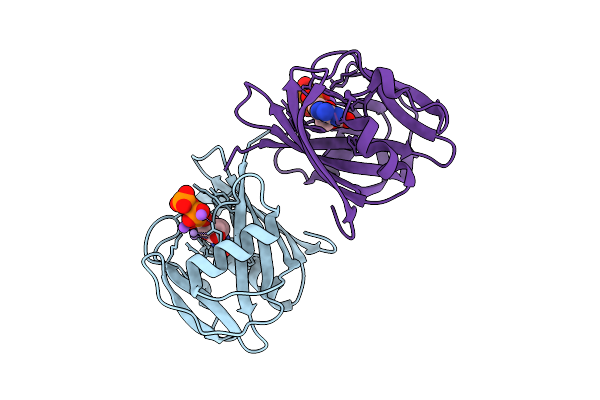
2-Oxo-Datp Hydrolysis In Human Mth1(G2K Mutant) Crystal Using Mn2+: The Es Complex
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: HYDROLASE Ligands: 6U4, NA |
 |
2-Oxo-Datp Hydrolysis In Human Mth1(G2K Mutant) Crystal Using Mn2+: The Es-2M Complex
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: HYDROLASE Ligands: 6U4, MN, NA |
 |
2-Oxo-Datp Hydrolysis In Human Mth1(G2K Mutant) Crystal Using Mn2+: The Es/Ep-3M Complex
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: HYDROLASE Ligands: 6U4, IGU, MN |
 |
2-Oxo-Datp Hydrolysis In Human Mth1(G2K Mutant) Crystal Using Mn2+: The Ep-M Complex
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: HYDROLASE Ligands: IGU, MN |
 |
Neutron And X-Ray Joint Refined Structure Of A Copper-Containing Nitrite Reductase (C135A Mutant) In Complex With Nitrite
Organism: Geobacillus thermodenitrificans ng80-2
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Release Date: 2025-06-11 Classification: OXIDOREDUCTASE Ligands: MPD, MRD, NO2, CU, DOD |
 |
Neutron And X-Ray Joint Refined Structure Of A Copper-Containing Nitrite Reductase (C135A Mutant) In Complex With Formate
Organism: Geobacillus thermodenitrificans (strain ng80-2)
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Release Date: 2025-06-11 Classification: OXIDOREDUCTASE Ligands: CU, FMT |
 |
D98N Mutant Of A Copper-Containing Nitrite Reductase From Geobacillus Thermodenitrificans
Organism: Geobacillus thermodenitrificans (strain ng80-2)
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: OXIDOREDUCTASE Ligands: CU, MPD, ACY, CL, NA |
 |
Structure Of A Copper-Containing Nitrite Reductase (D98N/G136A Mutant) From Geobacillus Thermodenitrificans
Organism: Geobacillus thermodenitrificans (strain ng80-2)
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: OXIDOREDUCTASE Ligands: CU, CL, MPD |
 |
D98N/C135A Mutant Of A Copper-Containing Nitrite Reductase In Complex With Nitrite
Organism: Geobacillus thermodenitrificans (strain ng80-2)
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: OXIDOREDUCTASE Ligands: CU, MPD, NO2 |
 |
Structure Of A D98N/C135A/G136A Mutant Copper-Containing Nitrite Reductase In Complex With Nitrite
Organism: Geobacillus thermodenitrificans (strain ng80-2)
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: OXIDOREDUCTASE Ligands: CU, MPD, NO2, CL |

