Search Count: 12
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-08-14 Classification: SUGAR BINDING PROTEIN Ligands: SO4 |
 |
Crystal And Solution Structures Of The Bifunctional Enzyme (Aldolase/Aldehyde Dehydrogenase) From Thermomonospora Curvata, Reveal A Cofactor-Binding Domain Motion During Nad+ And Coa Accommodation Whithin The Shared Cofactor-Binding Site
Organism: Thermomonospora curvata
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2013-09-04 Classification: OXIDOREDUCTASE Ligands: SO4, PYR, CL, MG, GOL, PEG, NAD, FOR |
 |
Crystal And Solution Structures Of The Bifunctional Enzyme (Aldolase/Aldehyde Dehydrogenase) From Thermomonospora Curvata, Reveal A Cofactor-Binding Domain Motion During Nad+ And Coa Accommodation Whithin The Shared Cofactor-Binding Site
Organism: Thermomonospora curvata
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2013-09-04 Classification: Lyase/OXIDOREDUCTASE Ligands: PYR, NA, MG, SO4, GOL, COA |
 |
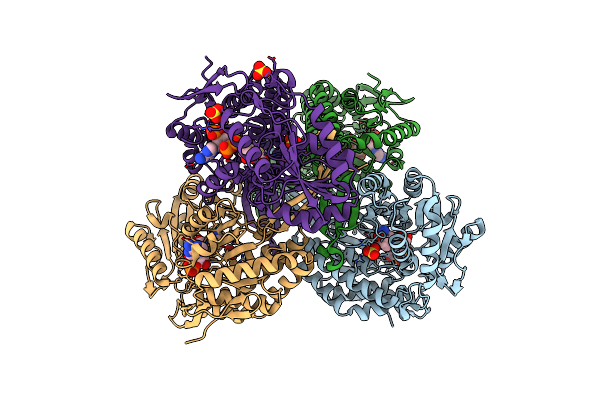
Thioacylenzyme Intermediate Of Gapn From S. Mutans, New Data Integration And Refinement.
Organism: Streptococcus mutans
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2007-12-25 Classification: OXIDOREDUCTASE Ligands: G3H, NAP |
 |
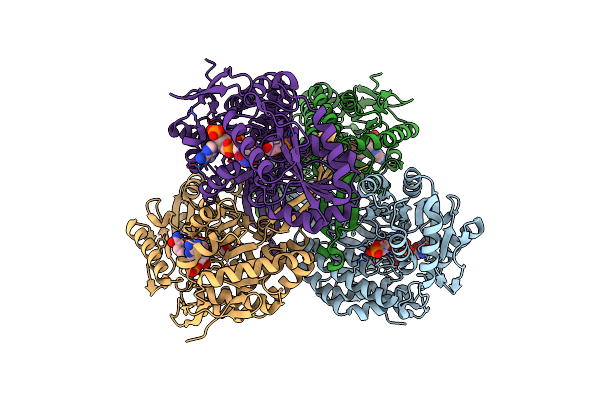
Organism: Streptococcus mutans
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2007-09-18 Classification: OXIDOREDUCTASE Ligands: SO4, NAP |
 |
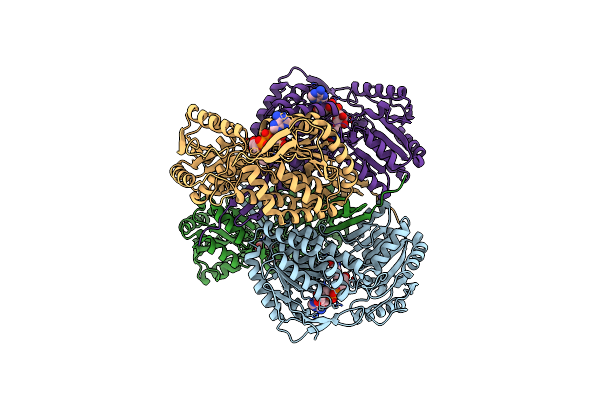
Crystal Structure Of Thioacylenzyme Intermediate Of An Nadp Dependent Aldehyde Dehydrogenase
Organism: Streptococcus mutans
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2006-05-02 Classification: OXIDOREDUCTASE Ligands: NAP, G3H |
 |
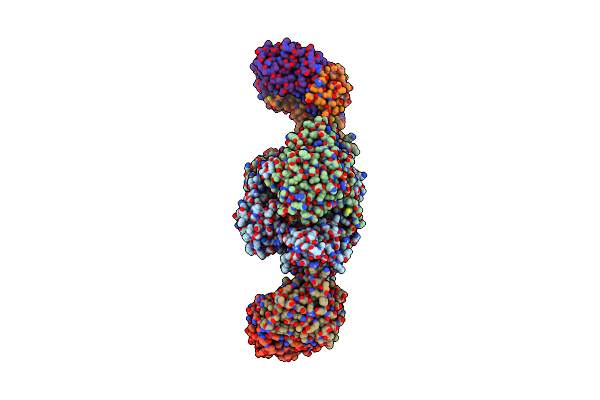
Crystal Structure Of Methylmalonate Semialdehyde Dehydrogenase From Bacillus Subtilis
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2006-01-03 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
Ternary Complex Between Trimethylamine Dehydrogenase And Electron Transferring Flavoprotein
Organism: Methylophilus methylotrophus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2003-02-06 Classification: ELECTRON TRANSPORT Ligands: FMN, ADP, SF4, AMP |
 |
Ternary Complex Between Trimethylamine Dehydrogenase And Electron Transferring Flavoprotein
Organism: Methylophilus methylotrophus
Method: X-RAY DIFFRACTION Resolution:3.70 Å Release Date: 2003-02-06 Classification: ELECTRON TRANSPORT Ligands: FMN, ADP, SF4, AMP |
 |
Structure Of Electron Transferring Flavoprotein For Methylophilus Methylotrophus.
Organism: Methylophilus methylotrophus
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2003-02-06 Classification: ELECTRON TRANSFER Ligands: AMP, FAD |
 |
Structure Of Electron Transferring Flavoprotein From Methylophilus Methylotrophus, Recognition Loop Removed By Limited Proteolysis
Organism: Methylophilus methylotrophus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2003-02-06 Classification: ELECTRON TRANSFER Ligands: AMP, FAD |
 |
Three-Dimensional Structure Of D-Glyceraldehyde-3-Phosphate Dehydrogenase From The Hyperthermophilic Archaeon Methanothermus Fervidus
Organism: Methanothermus fervidus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2000-03-29 Classification: OXIDOREDUCTASE Ligands: SO4, NAP |

