Search Count: 24
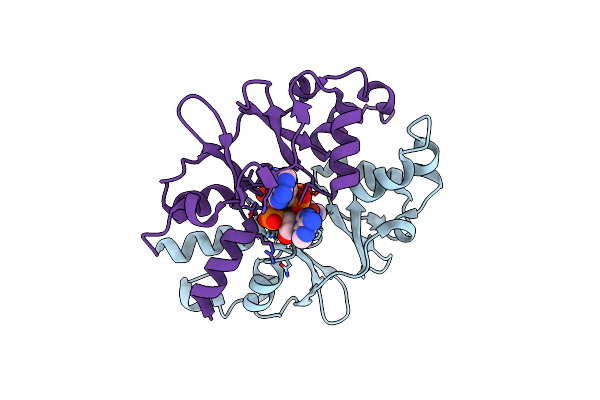 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2023-07-19 Classification: SIGNALING PROTEIN Ligands: 2BA |
 |
Organism: Salmonella
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2022-11-23 Classification: TOXIN Ligands: CL, NA |
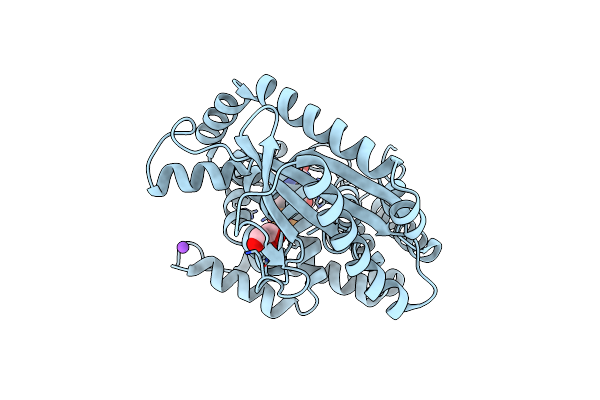 |
Structure Of The N-Terminal Catalytic Region Of T. Thermophilus Rel Bound To Ppgpp
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2020-07-08 Classification: TRANSFERASE Ligands: G4P, GOL, MN, MG, CL, NA |
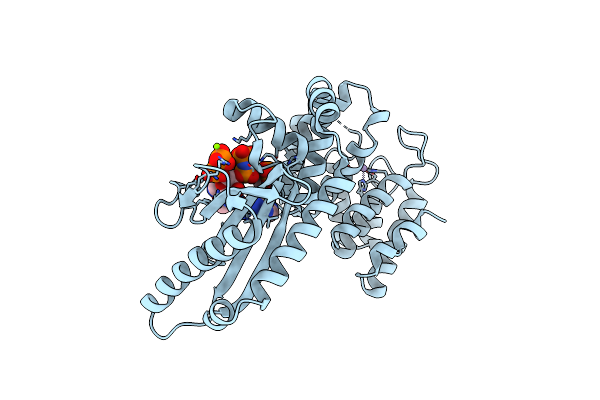 |
Structure Of The Catalytic Domain Of T. Thermophilus Rel In Complex With Amp And Ppgpp
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2020-07-08 Classification: TRANSFERASE Ligands: MN, AMP, GN3, TMO, MG |
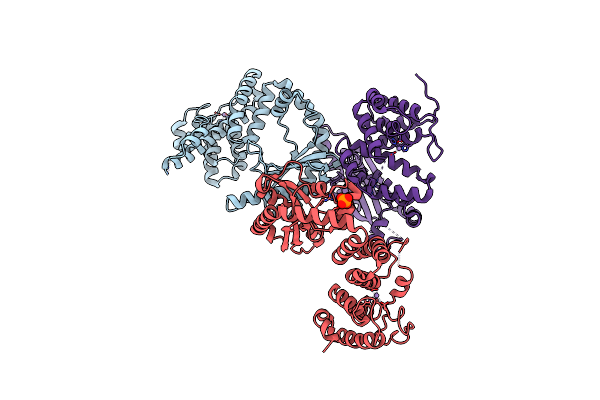 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.96 Å Release Date: 2020-07-08 Classification: TRANSFERASE Ligands: MN, CL, PO4, NA |
 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-01-30 Classification: ANTITOXIN |
 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:3.80 Å Release Date: 2019-01-30 Classification: ANTITOXIN |
 |
Organism: Pseudomonas putida, Pseudomonas putida (strain atcc 47054 / dsm 6125 / ncimb 11950 / kt2440)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2018-07-11 Classification: TOXIN Ligands: SO4, PEG |
 |
Structure Of Phosphorylated Translation Elongation Factor Ef-Tu From E. Coli
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2017-12-20 Classification: HYDROLASE Ligands: MG, GDP |
 |
Organism: Escherichia coli hs
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2017-12-20 Classification: HYDROLASE Ligands: MG, GDP, ACT, EPE, BME, CL |
 |
Organism: Escherichia coli o9:h4 (strain hs)
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2017-12-20 Classification: HYDROLASE Ligands: MG, GDP |
 |
Organism: Enterobacteria phage p1
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2016-04-20 Classification: TRANSCRIPTION Ligands: ACT |
 |
Antitoxin Phd From Phage P1 In Complex With Its Operator Dna Inverted Repeat
Organism: Enterobacteria phage p1
Method: X-RAY DIFFRACTION Resolution:3.17 Å Release Date: 2016-04-20 Classification: TRANSCRIPTION |
 |
Antitoxin Phd From Phage P1 In Complex With Its Operator Dna Inverted Repeat In A Monoclinic Space Group
Organism: Enterobacteria phage p1
Method: X-RAY DIFFRACTION Resolution:3.88 Å Release Date: 2016-04-20 Classification: TRANSCRIPTION |
 |
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2014-05-28 Classification: HYDROLASE |
 |
Structural And Biophysical Characterization Of The Mrna Interferase Samazf From Staphylococcus Aureus.
Organism: Staphylococcus aureus
Method: SOLUTION NMR Release Date: 2014-05-21 Classification: HYDROLASE |
 |
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2014-05-21 Classification: HYDROLASE |
 |
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-05-21 Classification: HYDROLASE |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2011-06-15 Classification: IMMUNE SYSTEM Ligands: PE4 |
 |
Crystal Structure Of Recombinant Kunitz Type Serine Protease Inhibitor-1 From The Caribbean Sea Anemone Stichodactyla Helianthus In Complex With Bovine Pancreatic Trypsin
Organism: Stichodactyla helianthus, Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2011-03-16 Classification: Hydrolase/Hydrolase inhibitor Ligands: PO4 |

