Search Count: 32
 |
Organism: Phage metagenome
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2025-09-17 Classification: VIRAL PROTEIN Ligands: OJC |
 |
Organism: Phage metagenome
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2025-07-30 Classification: VIRAL PROTEIN Ligands: OJC |
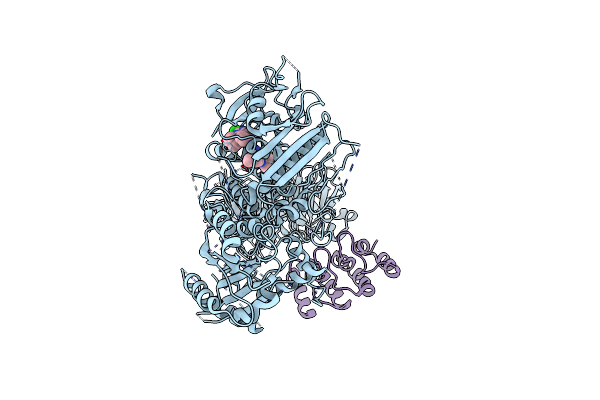 |
Structure Of The C-Terminal Half Of Lrrk2 Bound To Rn277 (Type-Ii Inhibitor)
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Resolution:3.35 Å Release Date: 2025-06-18 Classification: PROTEIN BINDING Ligands: A1A7Q |
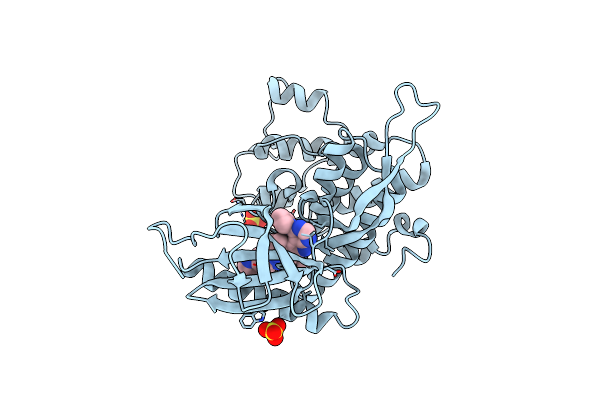 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2024-09-11 Classification: TRANSFERASE Ligands: SO4, A1H74 |
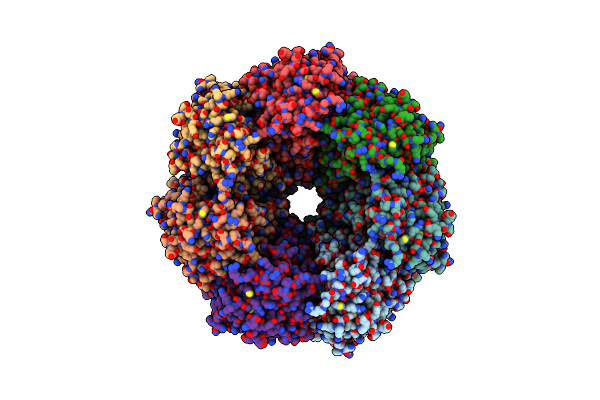 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-02-01 Classification: IMMUNE SYSTEM |
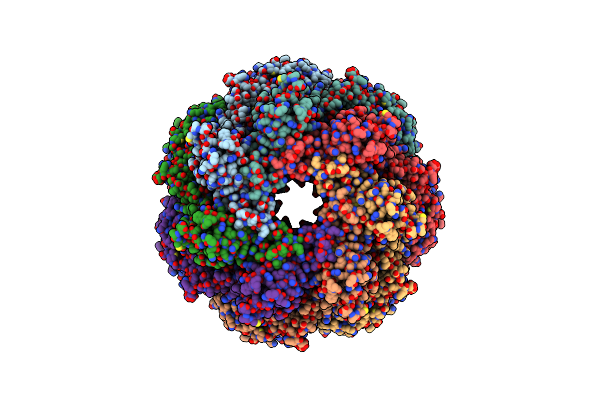 |
Organism: Streptococcus suis
Method: ELECTRON MICROSCOPY Release Date: 2023-02-01 Classification: IMMUNE SYSTEM |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-02-01 Classification: IMMUNE SYSTEM Ligands: ZN |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-02-01 Classification: IMMUNE SYSTEM Ligands: ZN |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2022-03-09 Classification: UNKNOWN FUNCTION |
 |
 |
Dimeric Photosystem I Of A Temperature Sensitive Mutant Chlamydomonas Reinhardtii
Organism: Chlamydomonas reinhardtii
Method: ELECTRON MICROSCOPY Release Date: 2021-12-15 Classification: PHOTOSYNTHESIS |
 |
 |
Structure Of Yersinia Aleksiciae Cap15 Cyclic Dinucleotide Receptor, Crystal Form 1
Organism: Yersinia aleksiciae
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2021-11-17 Classification: IMMUNE SYSTEM |
 |
Structure Of Yersinia Aleksiciae Cap15 Cyclic Dinucleotide Receptor, Crystal Form 2
Organism: Yersinia aleksiciae
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2021-11-17 Classification: IMMUNE SYSTEM |
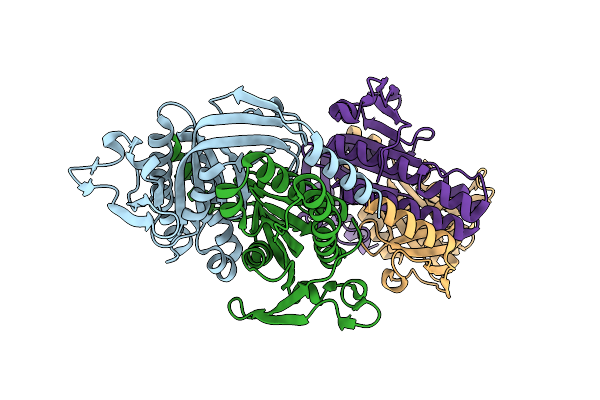 |
Crystal Structure Of A Bacterial Cyclic Ump Synthase From Burkholderia Cepacia Lk29
Organism: Burkholderia cepacia
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2021-10-13 Classification: TRANSFERASE |
 |
Organism: Plasmodium falciparum (isolate 3d7), Plasmodium falciparum
Method: ELECTRON MICROSCOPY Release Date: 2021-09-22 Classification: HYDROLASE Ligands: BO2 |
 |
Organism: Plasmodium falciparum (isolate 3d7)
Method: ELECTRON MICROSCOPY Release Date: 2021-09-22 Classification: HYDROLASE Ligands: YHD |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-09-22 Classification: HYDROLASE Ligands: YHD |
 |
Organism: Pisum sativum
Method: ELECTRON MICROSCOPY Release Date: 2021-06-16 Classification: PHOTOSYNTHESIS Ligands: CL0, CLA, PQN, SF4, BCR, LHG, LMT, LMG, CA, DGD, LUT, CHL, XAT, 3PH, CU |
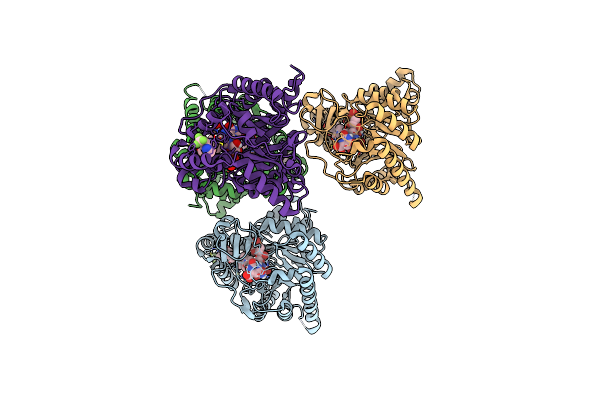 |
Crystal Structure Of Plasmodium Falciparum Dihydroorotate Dehydrogenase Bound With Inhibitor Dsm589 (Ethyl 3-Methyl-4-((4-(Trifluoromethyl)Benzo[D]Oxazol-7-Yl)Methyl)-1H-Pyrrole-2-Carboxylate)
Organism: Plasmodium falciparum (isolate 3d7)
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2021-05-19 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: XAJ, FMN, ORO |

