Search Count: 49
 |
Organism: Oryza sativa subsp. japonica
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: PLANT PROTEIN |
 |
Organism: Rhizopus arrhizus, Oryza sativa subsp. japonica
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: PROTEIN BINDING |
 |
Crystal Structure Of Bacteroides Thetaiotaomicron Gh84 O-Glcnacase D243N Mutant
Organism: Bacteroides thetaiotaomicron vpi-5482
Method: X-RAY DIFFRACTION Release Date: 2025-04-23 Classification: HYDROLASE Ligands: CA |
 |
Crystal Structure Of Chitin Oligosaccharide Binding Protein From Vibrio Cholera.
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-01-03 Classification: SUGAR BINDING PROTEIN Ligands: MG |
 |
Crystal Structure Of Chitin Oligosaccharide Binding Protein From Vibrio Cholera In Complex With Chitotriose.
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:1.22 Å Release Date: 2024-01-03 Classification: SUGAR BINDING PROTEIN Ligands: MG |
 |
Organism: Staphylococcus epidermidis
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2019-01-23 Classification: RNA BINDING PROTEIN |
 |
Organism: Thermoplasma volcanium (strain atcc 51530 / dsm 4299 / jcm 9571 / nbrc 15438 / gss1)
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2019-01-23 Classification: RNA BINDING PROTEIN |
 |
Organism: Escherichia coli, Enterobacteria phage qbeta
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2014-08-27 Classification: TRANSLATION/TRANSFERASE Ligands: SO4 |
 |
Crystal Structure Of Nadph-Dependent 3-Quinuclidinone Reductase From Rhodotorula Rubra
Organism: Rhodotorula mucilaginosa
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2014-08-13 Classification: OXIDOREDUCTASE Ligands: NDP |
 |
Crystal Structure Of 3-Quinuclidinone Reductase From Agrobacterium Tumefaciens
Organism: Agrobacterium tumefaciens
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2014-06-25 Classification: OXIDOREDUCTASE Ligands: NAD, ACY, EDO |
 |
Organism: Aquifex aeolicus
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2014-01-01 Classification: TRANSFERASE Ligands: SO4 |
 |
Organism: Aquifex aeolicus (strain vf5), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:4.01 Å Release Date: 2014-01-01 Classification: TRANSFERASE Ligands: SO4 |
 |
Organism: Synthetic construct, Aquifex aeolicus (strain vf5), Thermotoga maritima msb8
Method: X-RAY DIFFRACTION Resolution:3.62 Å Release Date: 2014-01-01 Classification: Transferase/RNA |
 |
Organism: Synthetic construct, Aquifex aeolicus (strain vf5), Thermotoga maritima msb8
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2014-01-01 Classification: Transferase/RNA Ligands: CTP, POP, MG |
 |
Organism: Synthetic construct, Aquifex aeolicus (strain vf5), Thermotoga maritima msb8
Method: X-RAY DIFFRACTION Resolution:3.31 Å Release Date: 2014-01-01 Classification: Transferase/RNA Ligands: SO4 |
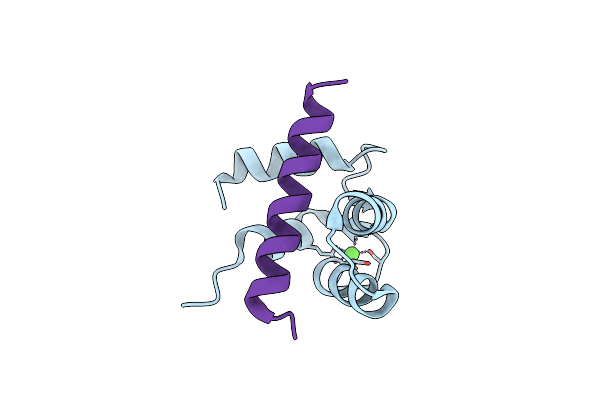 |
Crystal Structure Of The Ca2+-Saturated C-Terminal Domain Of Akazara Scallop Troponin C In Complex With A Troponin I Fragment
Organism: Chlamys nipponensis akazara
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2013-01-23 Classification: CONTRACTILE PROTEIN Ligands: CA |
 |
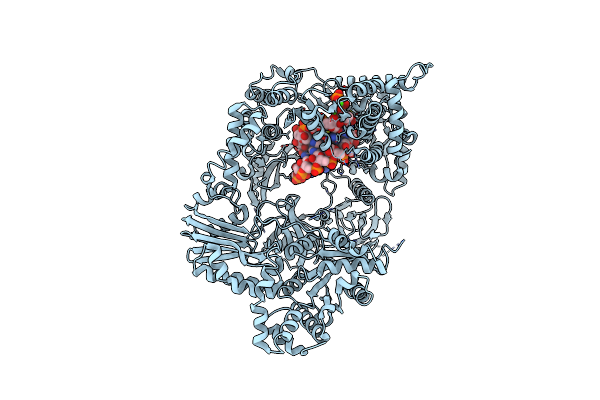
Organism: Escherichia coli o157:h7, Escherichia phage qbeta
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2012-08-08 Classification: TRANSLATION, TRANSFERASE/RNA Ligands: ATP, CA |
 |
Organism: Escherichia coli o157:h7, Escherichia phage qbeta
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2012-08-08 Classification: TRANSLATION, TRANSFERASE/RNA Ligands: CH1, CA |
 |
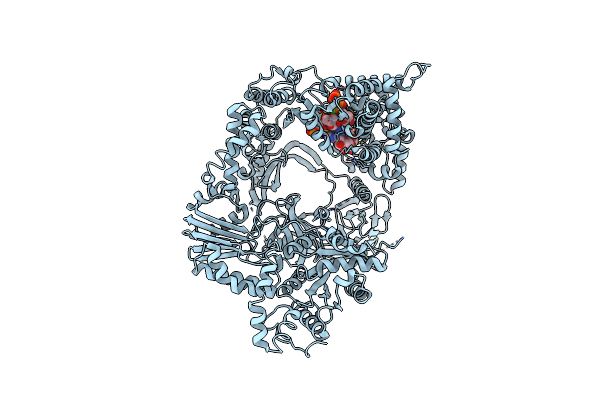
Organism: Escherichia coli o157:h7, Escherichia phage qbeta
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2012-08-08 Classification: TRANSLATION, TRANSFERASE/RNA Ligands: GTP, CA |
 |
Organism: Escherichia coli o157:h7, Synthetic construct, Escherichia phage qbeta
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2012-01-18 Classification: TRANSLATION, TRANSFERASE/RNA Ligands: GH3, CA |

