Search Count: 19
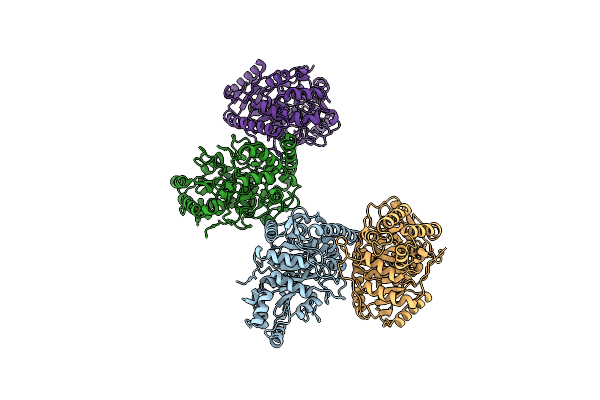 |
Organism: Pseudonocardia acaciae
Method: X-RAY DIFFRACTION Resolution:3.02 Å Release Date: 2024-10-09 Classification: HYDROLASE |
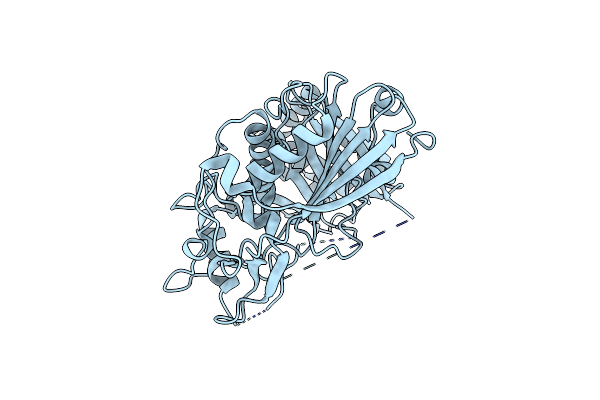 |
Organism: Triticum aestivum
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2021-11-17 Classification: TRANSFERASE |
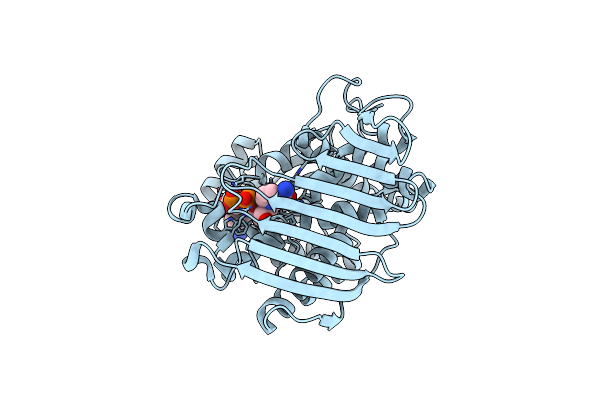 |
Crystal Structure Of Inositol Dehydrogenase Homolog Complexed With Nad+ From Azotobacter Vinelandii
Organism: Azotobacter vinelandii (strain dj / atcc baa-1303)
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2021-09-29 Classification: OXIDOREDUCTASE Ligands: NAD |
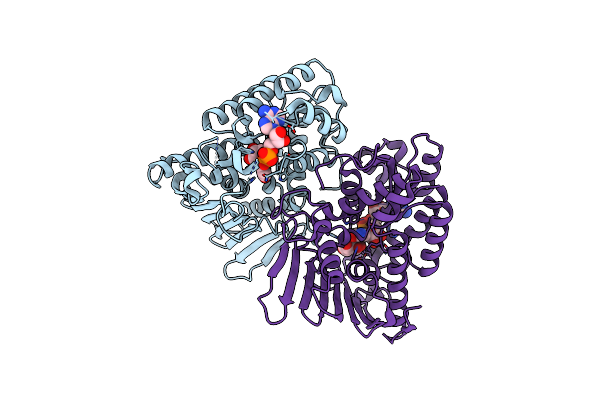 |
Crystal Structure Of Inositol Dehydrogenase Homolog Complexed With Nadh And Myo-Inositol From Azotobacter Vinelandii
Organism: Azotobacter vinelandii (strain dj / atcc baa-1303)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-09-29 Classification: OXIDOREDUCTASE Ligands: NAI, INS |
 |
Crystal Structure Of Barley Agmatine Coumaroyltransferase (Hvact), An N-Acyltransferase In Bahd Superfamily
Organism: Hordeum vulgare
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2020-12-16 Classification: TRANSFERASE |
 |
Crystal Structure Of A Substrate Binding Protein From Microbacterium Hydrocarbonoxydans
Organism: Microbacterium hydrocarbonoxydans
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2020-03-25 Classification: TRANSPORT PROTEIN |
 |
Crystal Structure Of A Substrate Binding Protein From Microbacterium Hydrocarbonoxydans Complexed With 4-Hydroxybenzoate Hydrazide
Organism: Microbacterium hydrocarbonoxydans
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-03-25 Classification: TRANSPORT PROTEIN Ligands: HDH |
 |
Crystal Structure Of The Substrate Binding Protein From Microbacterium Hydrocarbonoxydans Complexed With Propylparaben
Organism: Microbacterium hydrocarbonoxydans
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-03-25 Classification: TRANSPORT PROTEIN Ligands: 36M |
 |
Crystal Structure Of Scyllo-Inositol Dehydrogenase R178A Mutant, Apo-Form, From Paracoccus Laeviglucosivorans
Organism: Paracoccus laeviglucosivorans
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-12-25 Classification: OXIDOREDUCTASE Ligands: ACT |
 |
Crystal Structure Of Scyllo-Inositol Dehydrogenase R178A Mutant, Complexed With Nadh And L-Glucono-1,5-Lactone, From Paracoccus Laeviglucosivorans
Organism: Paracoccus laeviglucosivorans
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2019-12-25 Classification: OXIDOREDUCTASE Ligands: NAI, PG4, 8S0 |
 |
Crystal Structure Of Scyllo-Inositol Dehydrogenase R178A Mutant, Complexed With Nad And Myo-Inositol, From Paracoccus Laeviglucosivorans
Organism: Paracoccus laeviglucosivorans
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2019-12-25 Classification: OXIDOREDUCTASE Ligands: ACT, NAD, INS |
 |
Crystal Structure Of Aminoglycoside 7"-Phoshotransferase-Ia (Aph(7")-Ia/Hyg) From Streptomyces Hygroscopicus Complexed With Hygromycin B
Organism: Streptomyces hygroscopicus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2019-09-11 Classification: TRANSFERASE Ligands: HY0, NHE, FLC |
 |
Crystal Structure Of (S)-3-Hydroxybutyryl-Coenzymea Dehydrogenase From Clostridium Acetobutylicum Complexed With Nad+
Organism: Clostridium acetobutylicum (strain atcc 824 / dsm 792 / jcm 1419 / lmg 5710 / vkm b-1787)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-11-07 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
Crystal Structure Of (S)-3-Hydroxybutyryl-Coa Dehydrogenase From Clostridium Acetobutylicum, Apo Form
Organism: Clostridium acetobutylicum
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2018-11-07 Classification: OXIDOREDUCTASE |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2011-05-11 Classification: MEMBRANE PROTEIN Ligands: PO4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2010-09-22 Classification: MEMBRANE PROTEIN Ligands: BME, ZN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2010-09-22 Classification: MEMBRANE PROTEIN Ligands: SO4 |
 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2010-04-07 Classification: OXIDOREDUCTASE Ligands: NDP, MG, MPD |
 |
Crystal Structure Of Dxr From Thermooga Maritia, In Complex With Fosmidomycin And Nadph
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2010-03-16 Classification: OXIDOREDUCTASE Ligands: NDP, FOM, MG |

