Search Count: 17
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2022-01-19 Classification: DNA BINDING PROTEIN |
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2022-01-19 Classification: DNA BINDING PROTEIN |
 |
Crystal Structure Of A Far-Red Light-Absorbing Form Of Anpixjg2_Bv4 In Complex With Biliverdin
Organism: Nostoc sp.
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2019-04-17 Classification: SIGNALING PROTEIN Ligands: BLA, GOL |
 |
Xfel Structure Of Hen Egg-White Lysozyme Solved Using A Droplet Injector At Sacla
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2016-04-13 Classification: HYDROLASE Ligands: CL, NA |
 |
Organism: Delftia sp.
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2015-03-11 Classification: LYASE Ligands: PLP, MG |
 |
D-Threo-3-Hydroxyaspartate Dehydratase H351A Mutant Complexed With 2-Amino Maleic Acid
Organism: Delftia sp.
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-03-11 Classification: LYASE Ligands: PLP, MG, 2KZ |
 |
D-Threo-3-Hydroxyaspartate Dehydratase H351A Mutant Complexed With L-Erythro-3-Hydroxyaspartate
Organism: Delftia sp.
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2015-03-11 Classification: LYASE Ligands: PLP, MG, BH2, GOL |
 |
Crystal Structure Of A Potent Anti-Hiv Lectin Actinohivin In Complex With Alpha-1,2-Mannotriose
Organism: Actinomycete sp.
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2015-03-04 Classification: SUGAR BINDING PROTEIN |
 |
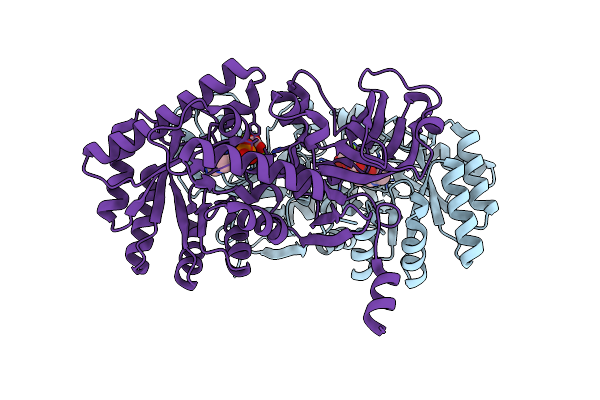
Organism: Delftia
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2015-01-28 Classification: LYASE Ligands: PLP, MG, GOL, CL |
 |
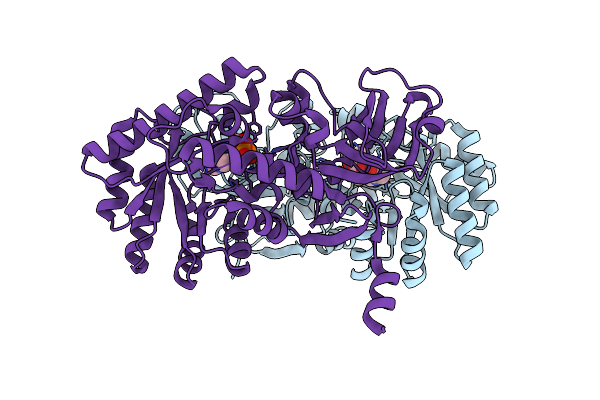
D-Threo-3-Hydroxyaspartate Dehydratase From Delftia Sp. Ht23 Complexed With D-Erythro-3-Hydroxyaspartate
Organism: Delftia
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2015-01-28 Classification: LYASE Ligands: PLP, MG, 999 |
 |
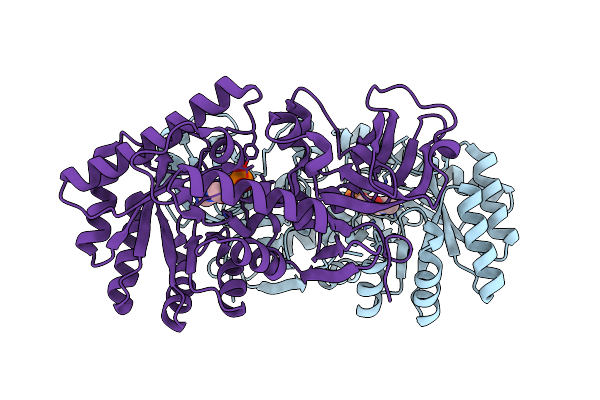
D-Threo-3-Hydroxyaspartate Dehydratase From Delftia Sp. Ht23 Complexed With D-Allothreonine
Organism: Delftia
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2015-01-28 Classification: LYASE Ligands: PLP, MG, 2TL |
 |
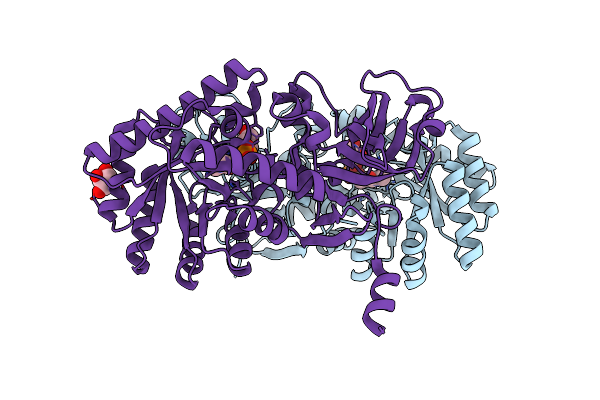
D-Threo-3-Hydroxyaspartate Dehydratase From Delftia Sp. Ht23 In The Metal-Free Form
Organism: Delftia
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2015-01-28 Classification: LYASE Ligands: PLP |
 |
Organism: Delftia
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2015-01-28 Classification: LYASE Ligands: PLP, GOL |
 |
Ha1 (Ha33) Subcomponent Of Botulinum Type C Progenitor Toxin Complexed With N-Acetylgalactosamine, Bound At Site Ii
Organism: Clostridium botulinum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2011-06-01 Classification: TOXIN Ligands: NGA |
 |
Ha1 (Ha33) Mutant F179I Of Botulinum Type C Progenitor Toxin Complexed With N-Acetylgalactosamine, Bound At Site Ii
Organism: Clostridium botulinum
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2011-06-01 Classification: TOXIN Ligands: NGA |
 |
Crystal Structure Of Streptomyces R61 Dd-Peptidase Complexed With A Novel Cephalosporin Analog Of Cell Wall Peptidoglycan
Organism: Streptomyces sp.
Method: X-RAY DIFFRACTION Resolution:1.17 Å Release Date: 2001-02-07 Classification: HYDROLASE Ligands: CEH |
 |
Protein-Dna Conformational Changes In The Crystal Structure Of A Lambda Cro-Operator Complex
Organism: Enterobacteria phage lambda
Method: X-RAY DIFFRACTION Resolution:3.90 Å Release Date: 1992-01-15 Classification: TRANSCRIPTION/DNA |

