Search Count: 25
 |
Organism: Colletotrichum orbiculare
Method: SOLUTION NMR Release Date: 2025-11-19 Classification: VIRAL PROTEIN |
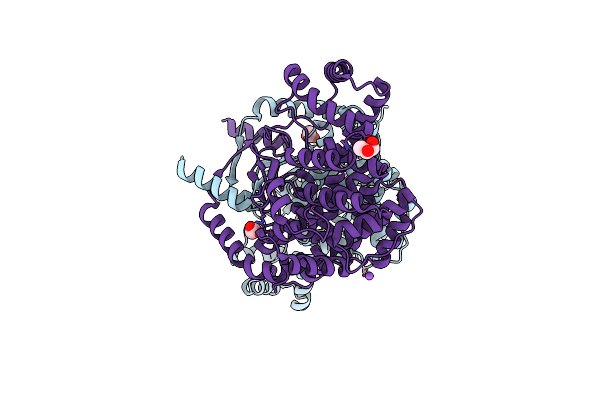 |
Crystal Structure Of Mitochondrial Citrate Synthase (Cit1) From Saccharomyces Cerevisiae
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2023-04-26 Classification: TRANSFERASE |
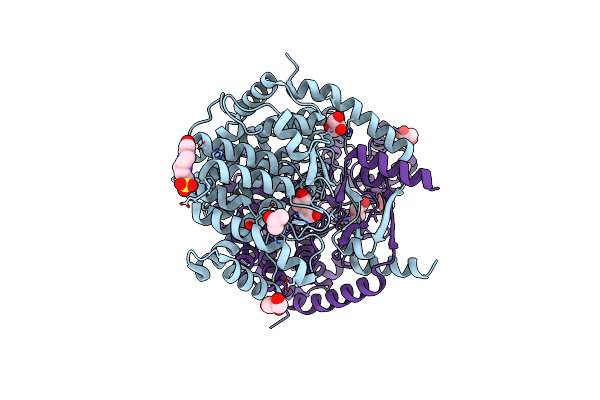 |
Crystal Structure Of Peroxisomal Citrate Synthase (Cit2) From Saccharomycescerevisiae
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2023-04-26 Classification: TRANSFERASE Ligands: CL, GOL, EPE, PEG |
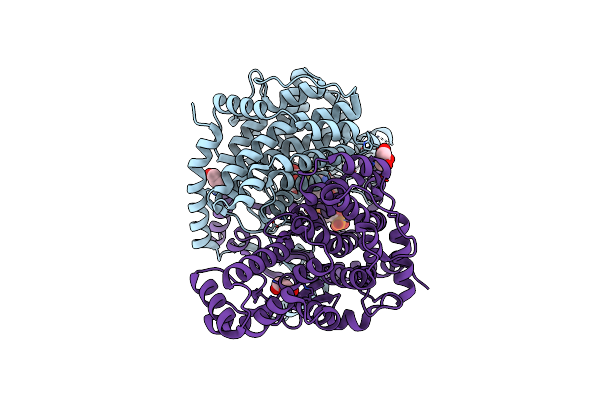 |
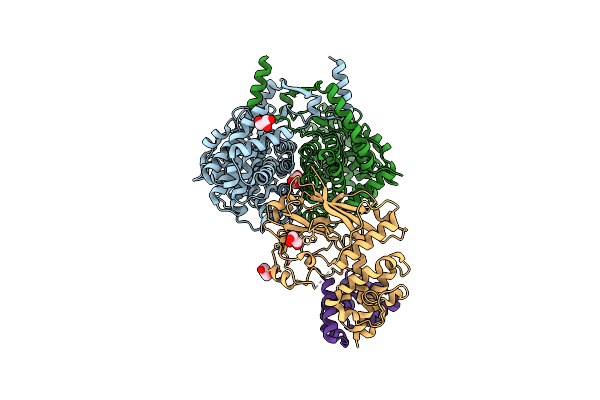
Crystal Structure Of Peroxisomal Citrate Synthase (Cit2) From Saccharomyces Cerevisiae In Complex With Oxaloacetate And Coenzyme-A
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2023-04-26 Classification: TRANSFERASE Ligands: COA, GOL, K, CL, OAA |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-04-26 Classification: TRANSFERASE/LIGASE Ligands: GOL |
 |
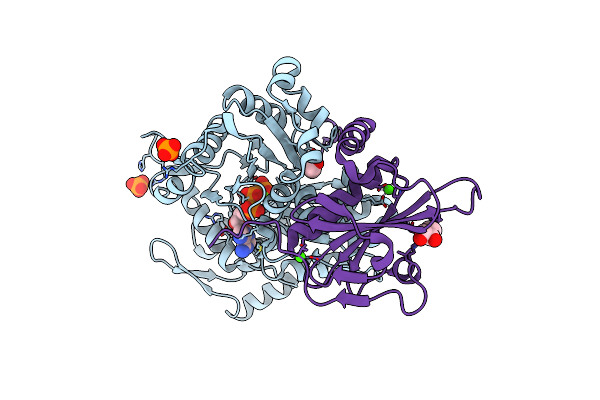
Crystal Structure Of F-Box Protein In The Ternary Complex With Adaptor Protein Skp1(Dl) And Its Substrate
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.53 Å Release Date: 2023-04-26 Classification: TRANSFERASE/LIGASE Ligands: EDO |
 |
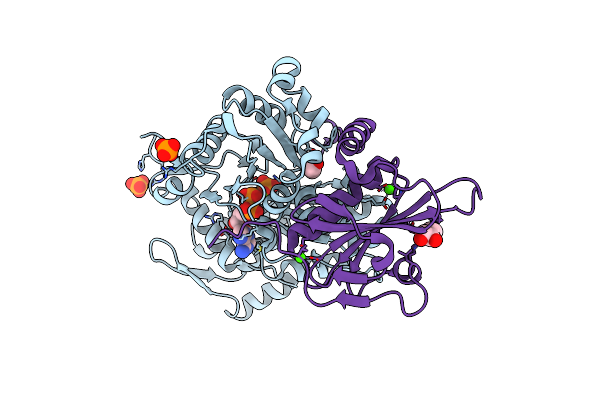
Organism: Physarum polycephalum, Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2022-10-26 Classification: CONTRACTILE PROTEIN Ligands: ANP, MG, PO4, EDO, CA |
 |
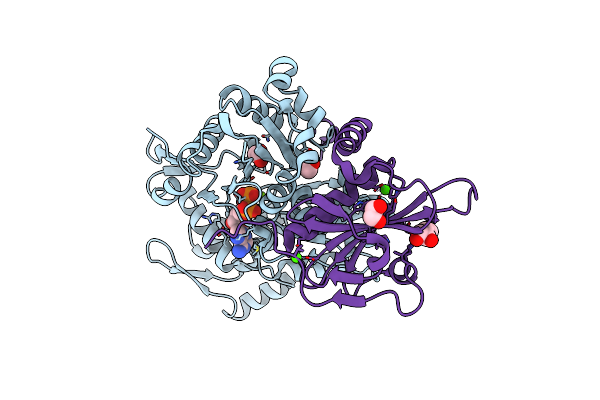
Organism: Physarum polycephalum, Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2022-10-26 Classification: CONTRACTILE PROTEIN Ligands: ADP, PO4, MG, EDO, CA |
 |
Organism: Physarum polycephalum, Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2022-10-26 Classification: CONTRACTILE PROTEIN Ligands: ADP, MG, EDO, CA |
 |
Organism: Physarum polycephalum, Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-10-26 Classification: CONTRACTILE PROTEIN Ligands: ATP, CA, ACT, NA, EDO |
 |
Organism: Physarum polycephalum, Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2022-10-26 Classification: CONTRACTILE PROTEIN Ligands: ADP, CA, ACT, NA, EDO, PO4 |
 |
Organism: Colletotrichum orbiculare (strain 104-t / atcc 96160 / cbs 514.97 / lars 414 / maff 240422)
Method: SOLUTION NMR Release Date: 2021-11-03 Classification: TOXIN |
 |
Structure Of Reaction Intermediate Of Cytochrome P450 No Reductase (P450Nor) Determined By Xfel
Organism: Fusarium oxysporum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-05-19 Classification: OXIDOREDUCTASE Ligands: HEM, NO, GOL |
 |
Sfx Structure Of Cytochrome P450Nor: A Complete Dark Data Without Pump Laser (Resting State)
Organism: Fusarium oxysporum
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-08-08 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
Structure Of Cytochrome P450Nor In No-Bound State: Damaged By Low-Dose (0.72 Mgy) X-Ray
Organism: Fusarium oxysporum
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2017-12-06 Classification: OXIDOREDUCTASE Ligands: HEM, NO, GOL |
 |
Structure Of Cytochrome P450Nor In No-Bound State: Damaged By High-Dose (5.7 Mgy) X-Ray
Organism: Fusarium oxysporum
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2017-12-06 Classification: OXIDOREDUCTASE Ligands: HEM, NO, GOL |
 |
Sf-Rox Structure Of Cytochrome P450Nor (No-Bound State) Determined At Sacla
Organism: Fusarium oxysporum
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2017-12-06 Classification: OXIDOREDUCTASE Ligands: HEM, NO, GOL |
 |
Time-Resolved Sfx Structure Of Cytochrome P450Nor: 20 Ms After Photo-Irradiation Of Caged No In The Presence Of Nadh (No-Bound State), Light Data
Organism: Fusarium oxysporum
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-12-06 Classification: OXIDOREDUCTASE Ligands: HEM, NO, GOL |
 |
Time-Resolved Sfx Structure Of Cytochrome P450Nor: Dark-2 Data In The Presence Of Nadh (Resting State)
Organism: Fusarium oxysporum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-12-06 Classification: OXIDOREDUCTASE Ligands: HEM, GOL |
 |
Time-Resolved Sfx Structure Of Cytochrome P450Nor : 20 Ms After Photo-Irradiation Of Caged No In The Absence Of Nadh (No-Bound State), Light Data
Organism: Fusarium oxysporum
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-12-06 Classification: OXIDOREDUCTASE Ligands: HEM, NO |

