Search Count: 18
 |
Crystal Structure Of A Highly Photostable And Bright Green Fluorescent Protein At Ph8.5
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2023-10-04 Classification: FLUORESCENT PROTEIN Ligands: CL |
 |
Crystal Structure Of A Highly Photostable And Bright Green Fluorescent Protein At Ph5.6
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-10-04 Classification: FLUORESCENT PROTEIN Ligands: CL |
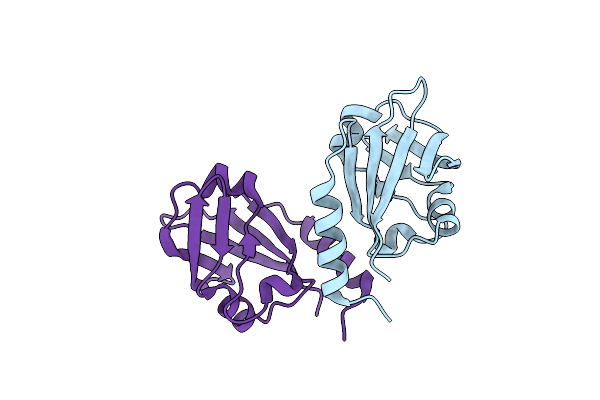 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2012-12-19 Classification: DNA BINDING PROTEIN Ligands: CL |
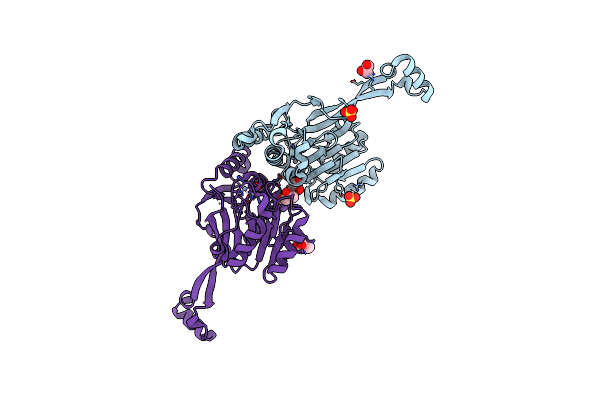 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2007-09-11 Classification: HYDROLASE Ligands: ZN, SO4, PGO |
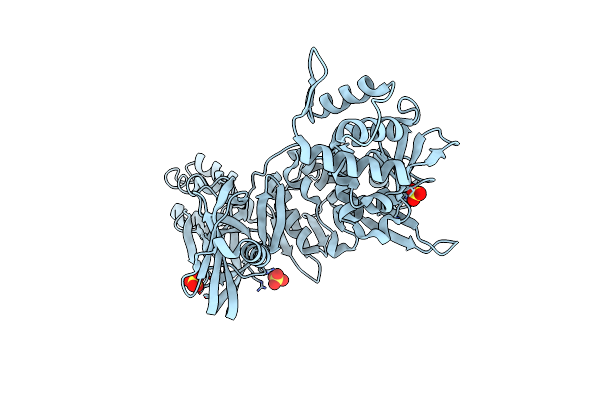 |
Organism: Thermococcus kodakarensis
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2006-04-18 Classification: HYDROLASE Ligands: SO4 |
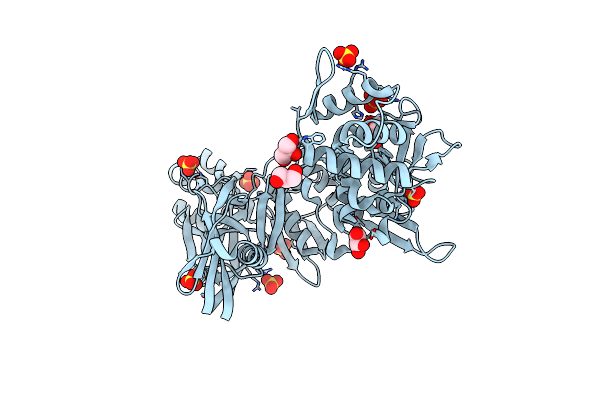 |
Organism: Thermococcus kodakarensis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2006-04-18 Classification: HYDROLASE Ligands: SO4, GOL |
 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2005-02-22 Classification: HYDROLASE Ligands: ZN |
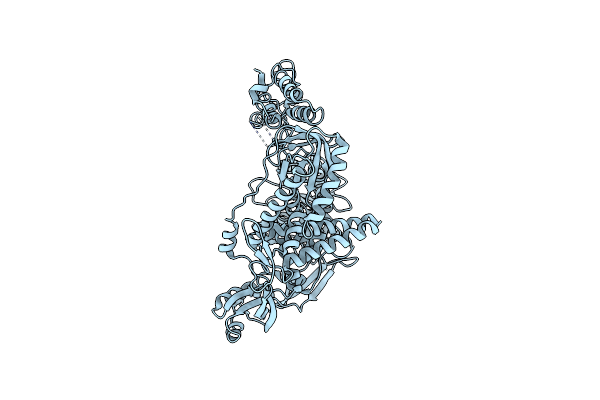 |
Crystal Structure Of Family B Dna Polymerase From Hyperthermophilic Archaeon Pyrococcus Kodakaraensis Kod1
Organism: Thermococcus kodakarensis
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2004-08-17 Classification: TRANSFERASE |
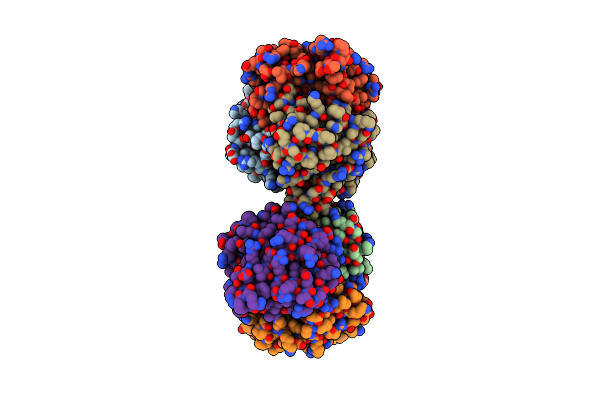 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2002-09-11 Classification: LYASE |
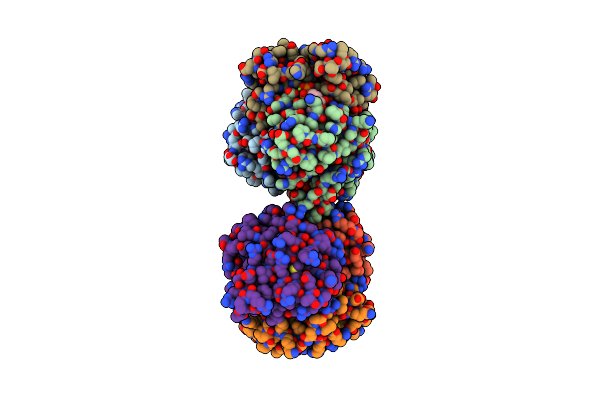 |
Structure Of 2C-Methyl-D-Erythritol-2,4-Cyclodiphosphate Synthase (Bound Form Cdp)
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2002-09-11 Classification: LYASE Ligands: MG, CDP |
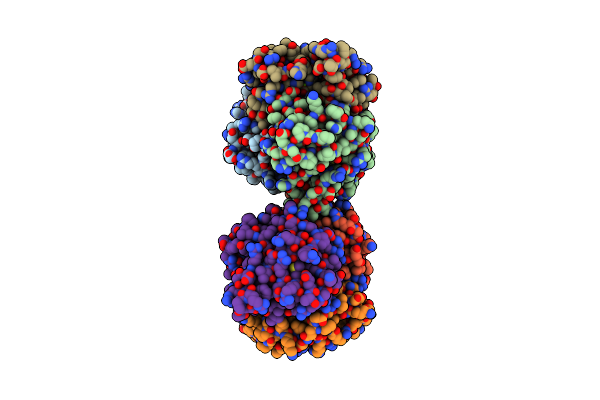 |
Structure Of 2C-Methyl-D-Erythritol-2,4-Cyclodiphosphate Synthase (Bound Form Mg Atoms)
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2002-09-11 Classification: LYASE Ligands: MG |
 |
Structure Of 2C-Methyl-D-Erythritol-2,4-Cyclodiphosphate Synthase (Bound Form Substrate)
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2002-09-11 Classification: LYASE Ligands: MG, C5P |
 |
Hydrolytic Haloalkane Dehalogenase Linb From Sphingomonas Paucimobilis Ut26 At 1.6 A Resolution
Organism: Sphingomonas paucimobilis
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2000-09-11 Classification: HYDROLASE |
 |
Hydrolytic Haloalkane Dehalogenase Linb From Sphingomonas Paucimobilis Ut26 With 1,3-Propanediol, A Product Of Debromidation Of Dibrompropane, At 2.0A Resolution
Organism: Sphingomonas paucimobilis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2000-09-11 Classification: HYDROLASE Ligands: BR, PDO |
 |
Crystal Structure Of O6-Methylguanine-Dna Methyltransferase From Hyperthermophilic Archaeon Pyrococcus Kodakaraensis Strain Kod1
Organism: Thermococcus kodakarensis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2000-01-07 Classification: TRANSFERASE Ligands: SO4 |
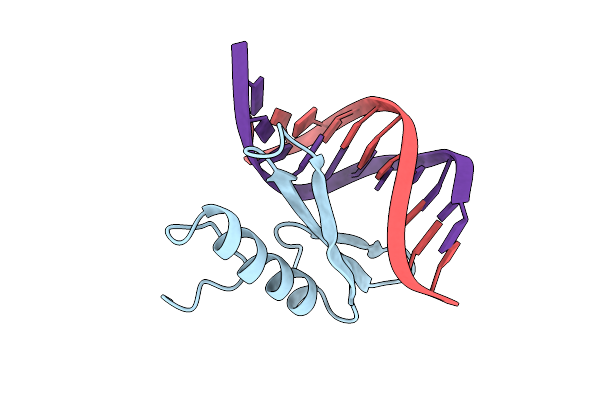 |
Solution Nmr Structure Of The Complex Of Gcc-Box Binding Domain Of Aterf1 And Gcc-Box Dna, Minimized Average Structure
Organism: Arabidopsis thaliana
Method: SOLUTION NMR Release Date: 1999-03-23 Classification: TRANSCRIPTION/DNA |
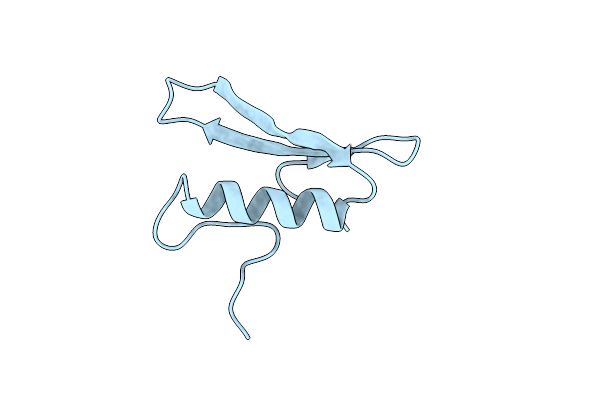 |
Solution Structure Of The Gcc-Box Binding Domain, Nmr, Minimized Mean Structure
Organism: Arabidopsis thaliana
Method: SOLUTION NMR Release Date: 1999-03-23 Classification: TRANSCRIPTION FACTOR |
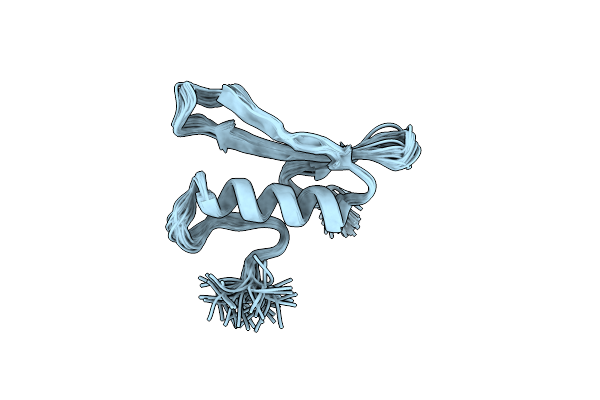 |
Organism: Arabidopsis thaliana
Method: SOLUTION NMR Release Date: 1999-03-23 Classification: TRANSCRIPTION FACTOR |

