Search Count: 18
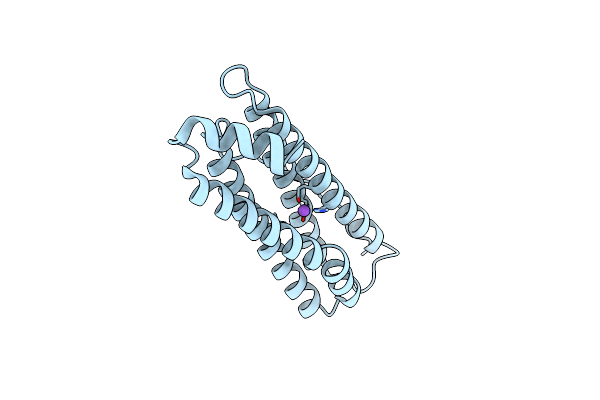 |
Single-Particle Cryo-Em Structure Of Mouse Apoferritin At 1.19 Angstrom Resolution (Dataset A)
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Resolution:1.19 Å Release Date: 2023-07-12 Classification: STRUCTURAL PROTEIN |
 |
Electron Crystallographic Structure Of Tia-1 Prion-Like Domain, A381T Mutant
Organism: Homo sapiens
Method: ELECTRON CRYSTALLOGRAPHY Resolution:0.95 Å Release Date: 2022-09-28 Classification: PROTEIN FIBRIL |
 |
Electron Crystallographic Structure Of Tia-1 Prion-Like Domain, Wild Type Sequence
Organism: Homo sapiens
Method: ELECTRON CRYSTALLOGRAPHY Resolution:1.76 Å Release Date: 2022-09-28 Classification: PROTEIN FIBRIL |
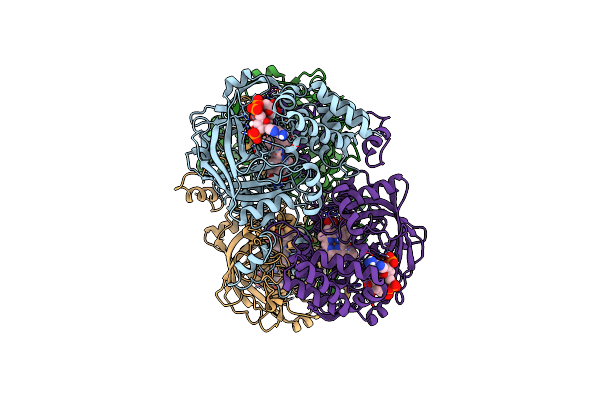 |
Electron Crystallographic Structure Of Catalase Using A Direct Electron Detector At 300 Kv
Organism: Bos taurus
Method: ELECTRON CRYSTALLOGRAPHY Resolution:3.20 Å Release Date: 2020-12-09 Classification: OXIDOREDUCTASE Ligands: HEM, NDP |
 |
Crystal Structure Of The S65T/F99S/M153T/V163A Variant Of Perdeuterated Gfp At Pd 8.5
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:0.90 Å Release Date: 2019-12-11 Classification: FLUORESCENT PROTEIN Ligands: DOD |
 |
Crystal Structure Of The S65T/F99S/M153T/V163A Variant Of Perdeuterated Gfp At Pd 7.0
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:0.80 Å Release Date: 2019-12-11 Classification: FLUORESCENT PROTEIN Ligands: DOD |
 |
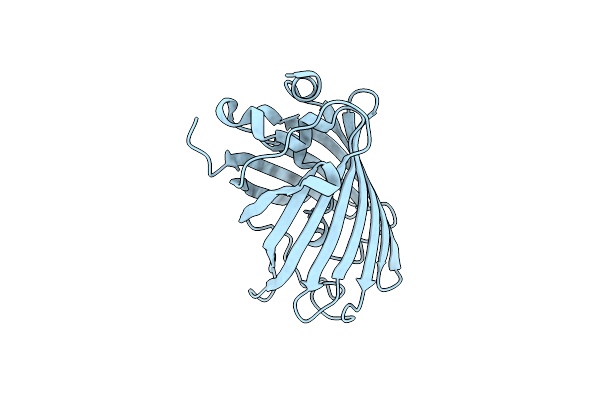
Crystal Structure Of The S65T/F99S/M153T/V163A Variant Of Non-Deuterated Gfp At Pd 8.5
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:0.85 Å Release Date: 2019-12-11 Classification: FLUORESCENT PROTEIN Ligands: DOD |
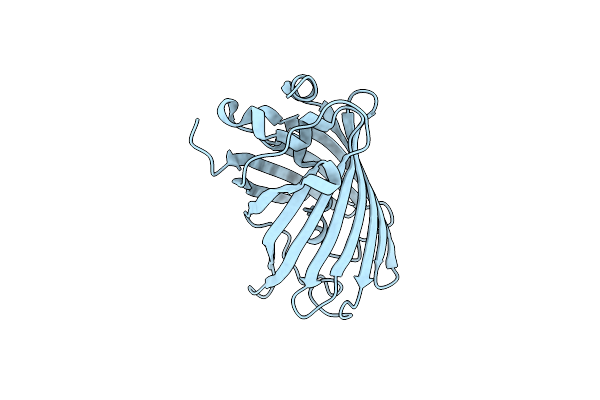
 |
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:0.94 Å Release Date: 2019-04-17 Classification: FLUORESCENT PROTEIN Ligands: CL |
 |
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:0.85 Å Release Date: 2019-04-17 Classification: FLUORESCENT PROTEIN |
 |
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:0.78 Å Release Date: 2019-04-17 Classification: FLUORESCENT PROTEIN Ligands: MG |
 |
Structure Of Nadh-Cytochrome B5 Reductase Refined With The Multipolar Atomic Model At 0.80 A
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:0.80 Å Release Date: 2017-04-05 Classification: OXIDOREDUCTASE Ligands: FAD, GOL |
 |
Structure Of Nadh-Cytochrome B5 Reductase Refined With The Multipolar Atomic Model At 0.78A
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:0.78 Å Release Date: 2017-04-05 Classification: OXIDOREDUCTASE Ligands: FAD, GOL |
 |
Crystal Structure Of Oxidation Intermediate (20 Min) Of Nadh-Cytochrome B5 Reductase From Pig Liver
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2013-07-17 Classification: OXIDOREDUCTASE Ligands: FAD, NAD |
 |
Crystal Structure Of Oxidation Intermediate (10 Min) Of Nadh-Cytochrome B5 Reductase From Pig Liver
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2013-07-17 Classification: OXIDOREDUCTASE Ligands: FAD, NAD |
 |
Crystal Structure Of Fully Reduced Form Of Nadh-Cytochrome B5 Reductase From Pig Liver
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2013-07-17 Classification: OXIDOREDUCTASE Ligands: FAD, NAD |
 |
Crystal Structure Of Oxidation Intermediate (1Min) Of Nadh-Cytochrome B5 Reductase From Pig Liver
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2013-07-17 Classification: OXIDOREDUCTASE Ligands: FAD, NAD |
 |
Crystal Structure Of Re-Oxidized Form (60 Min) Of Nadh-Cytochrome B5 Reductase From Pig Liver
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2013-07-17 Classification: OXIDOREDUCTASE Ligands: FAD, NAD |
 |
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:0.78 Å Release Date: 2013-07-17 Classification: OXIDOREDUCTASE Ligands: FAD, GOL |

