Search Count: 23
 |
Organism: Hepacivirus hominis, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RIBOSOME Ligands: ZN, MG |
 |
Structure Of The Human 40S Ribosome Complexed With Hcv Ires, Eif1A And Eif3
Organism: Homo sapiens, Hepacivirus hominis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RIBOSOME Ligands: ZN, MG |
 |
Structure Of The Hcv Ires-Dependent Pre-48S Translation Initiation Complex With Eif1A, Eif5B, And Eif3
Organism: Homo sapiens, Hepacivirus hominis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RIBOSOME Ligands: MG, GTP, ZN |
 |
Structure Of The Hcv Ires-Dependent 48S Translation Initiation Complex With Eif5B And Eif3
Organism: Homo sapiens, Hepacivirus hominis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RIBOSOME Ligands: ZN, MG, GTP |
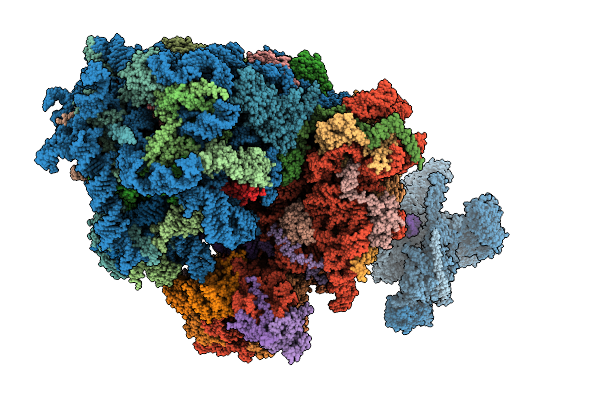 |
Cryo-Em Structure Of The Hcv Ires-Dependently Initiated Cmv-Stalled 80S Ribosome (Non-Rotated State) In Complexed With Eif3
Organism: Hepacivirus hominis, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RIBOSOME Ligands: MG, ZN |
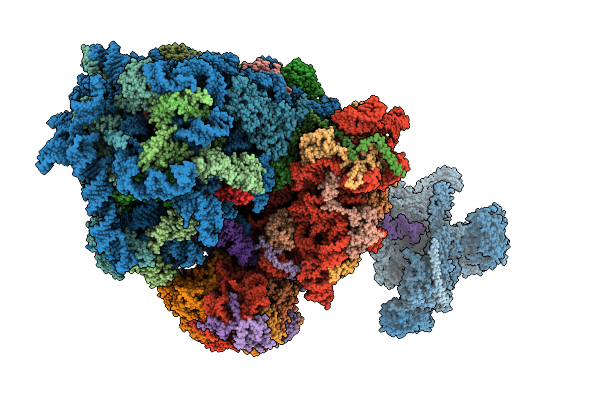 |
Cryo-Em Structure Of The Hcv Ires-Dependently Initiated Cmv-Stalled 80S Ribosome (Rotated State) In Complexed With Eif3
Organism: Hepacivirus hominis, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RIBOSOME Ligands: MG, ZN |
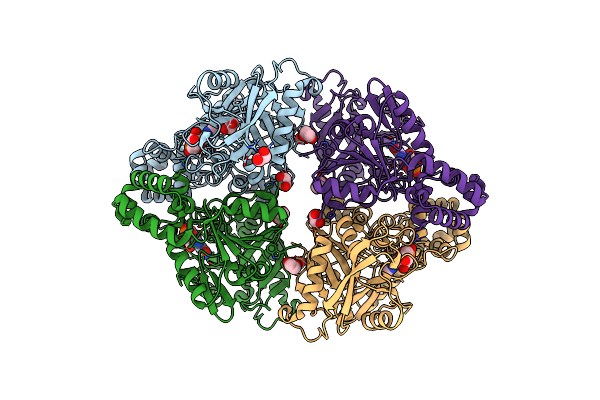 |
Crystal Structure Of Adenylosuccinate Synthetase, Pura, From Thermus Thermophilus
Organism: Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-04-08 Classification: LIGASE Ligands: IMP, EDO |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2019-05-29 Classification: RIBOSOME |
 |
Cryo-Em Structure Of The Cmv-Stalled Human 80S Ribosome With Hcv Ires (Structure Iii)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2019-05-29 Classification: RIBOSOME |
 |
Cryo-Em Structure Of The Hcv Ires Dependently Initiated Cmv-Stalled 80S Ribosome (Structure Iv)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2019-05-29 Classification: RIBOSOME |
 |
Crystal Structure Of The Human Eif4A1-Atp Analog-Roca-Polypurine Rna Complex
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-01-16 Classification: TRANSLATION/RNA Ligands: ANP, MG, RCG |
 |
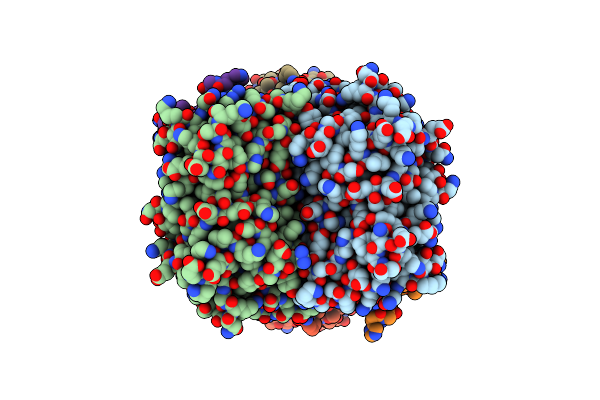
Crystal Structure Of Formyltetrahydrofolate Deformylase From Thermus Thermophilus Hb8
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.71 Å Release Date: 2014-01-08 Classification: HYDROLASE |
 |
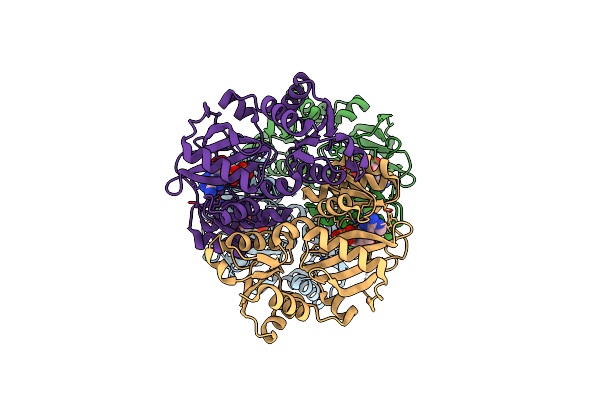
Organism: Sulfolobus tokodaii
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2013-02-27 Classification: LIGASE Ligands: ADP, GLU, MG, ZN, SO4 |
 |
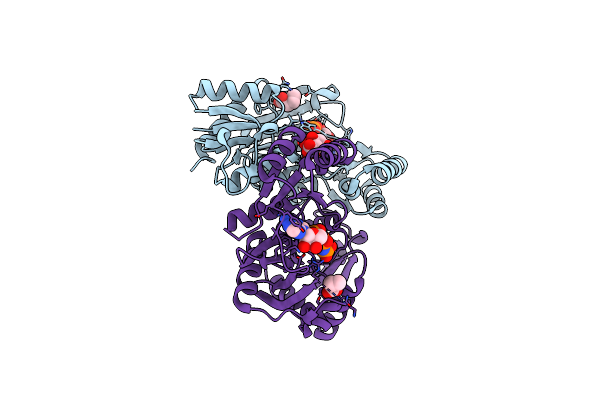
Organism: Sulfolobus tokodaii
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2013-02-27 Classification: LIGASE Ligands: ADP |
 |
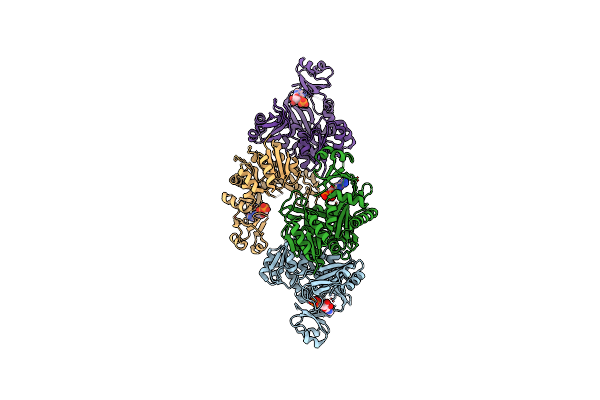
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2013-02-27 Classification: LIGASE Ligands: ANP, BUA, CIT |
 |
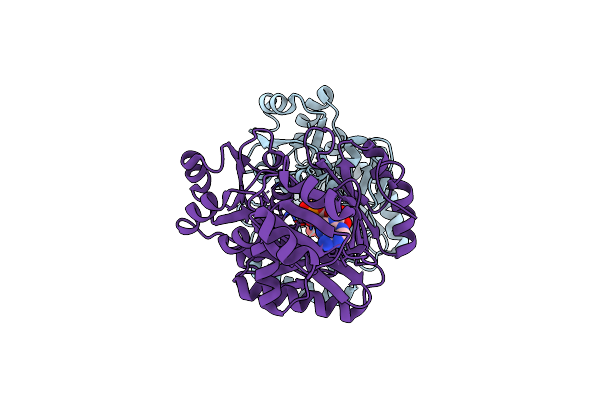
Crystal Structure Of N5-Carboxyaminoimidazole Ribonucleotide Synthetase From Thermotoga Maritima
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2012-04-25 Classification: LIGASE Ligands: ADP |
 |
Crystal Structure Of N5-Carboxyaminoimidazole Ribonucleotide Synthetase From Thermus Thermophilus Hb8
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2012-04-11 Classification: LIGASE Ligands: AMP, CL |
 |
Crystal Structure Of Glycinamide Ribonucleotide Transformylase 1 From Symbiobacterium Toebii
Organism: Symbiobacterium toebii
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2012-03-07 Classification: TRANSFERASE |
 |
Crystal Structure Of Glycinamide Ribonucleotide Transformylase 1 From Geobacillus Kaustophilus
Organism: Geobacillus kaustophilus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2012-03-07 Classification: TRANSFERASE Ligands: MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2009-09-01 Classification: TRANSLATION Ligands: SO4, CL |

