Search Count: 411
 |
Crystal Structure Of Human Apoptosis-Inducing Factor (Aif) Bound To The Fused N-Terminal Domain Of Chchd4
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-01-08 Classification: OXIDOREDUCTASE Ligands: FAD, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2024-11-06 Classification: RECOMBINATION Ligands: ADP, BEF, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2024-07-10 Classification: RNA BINDING PROTEIN |
 |
Organism: Alvinella pompejana
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2024-07-10 Classification: RNA BINDING PROTEIN Ligands: IMD, EDO |
 |
Crystal Structure Of Human Apoptosis-Inducing Factor (Aif) W196A Mutant Complexed With 6-Fluoroquinolin-4-Amine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2023-11-08 Classification: OXIDOREDUCTASE Ligands: FAD, QDI, EDO, CL |
 |
Crystal Structure Of Human Apoptosis-Inducing Factor (Aif) W196A Mutant Complexed With 6-Chloroquinolin-4-Amine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2023-11-08 Classification: OXIDOREDUCTASE Ligands: FAD, QD1, EDO |
 |
Crystal Structure Of Human Apoptosis-Inducing Factor (Aif) W196A Mutant Complexed With 7-Chloroquinolin-4-Amine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2023-11-08 Classification: OXIDOREDUCTASE Ligands: FAD, QBC, EDO |
 |
Crystal Structure Of Human Apoptosis-Inducing Factor (Aif) W196A Mutant Complexed With Quinolin-4-Amine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2023-11-08 Classification: OXIDOREDUCTASE Ligands: 1LM, EDO, FAD, SO4 |
 |
Crystal Structure Of Human Apoptosis-Inducing Factor (Aif) Complexed With 6-Fluoro-2-Methylquinolin-4-Amine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2023-11-08 Classification: OXIDOREDUCTASE Ligands: EDO, FAD, QEF, SO4 |
 |
Crystal Structure Of Human Apoptosis-Inducing Factor (Aif) Complexed With 8-Fluoro-2-Methylquinolin-4-Amine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-11-08 Classification: OXIDOREDUCTASE Ligands: QC6, EDO, FAD, SO4 |
 |
Crystal Structure Of Human Apoptosis-Inducing Factor (Aif) Complexed With 7-Chloroquinolin-4-Amine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2023-11-08 Classification: OXIDOREDUCTASE Ligands: QBC, FAD, EDO, SO4 |
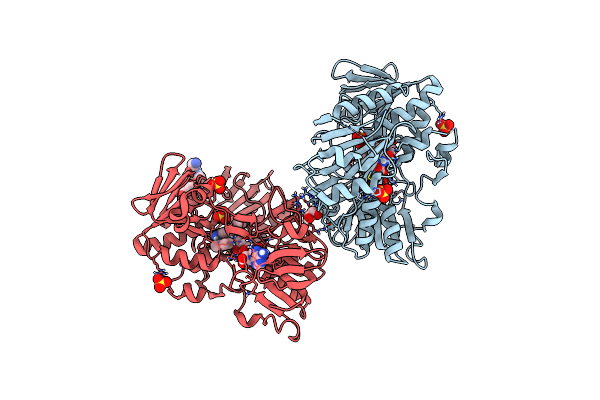 |
Crystal Structure Of Human Apoptosis-Inducing Factor (Aif) Complexed With 8-Methoxyquinolin-4-Amine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2023-11-08 Classification: OXIDOREDUCTASE Ligands: QF6, EDO, FAD, SO4 |
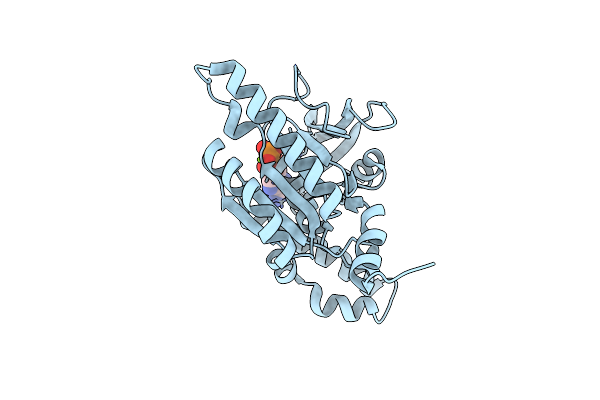 |
Organism: Alvinella pompejana
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-08-16 Classification: DNA BINDING PROTEIN Ligands: MG, ADP |
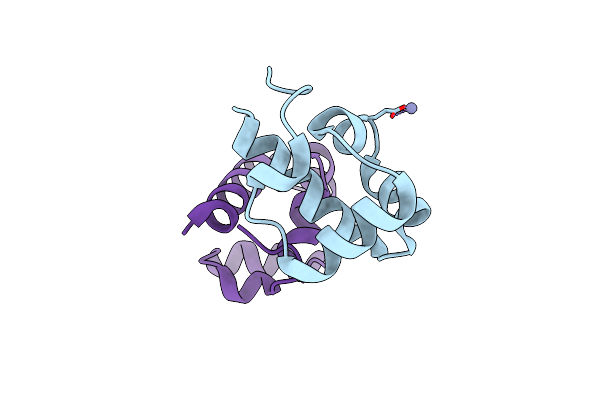 |
Organism: Alvinella pompejana
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-08-16 Classification: DNA BINDING PROTEIN Ligands: ZN |
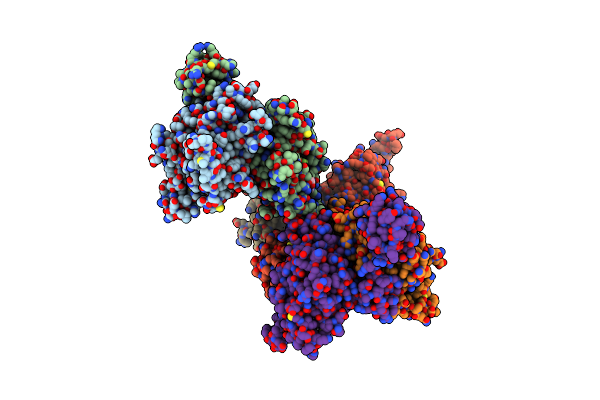 |
Organism: Alvinella pompejana
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2023-08-16 Classification: DNA BINDING PROTEIN Ligands: ADP, BEF |
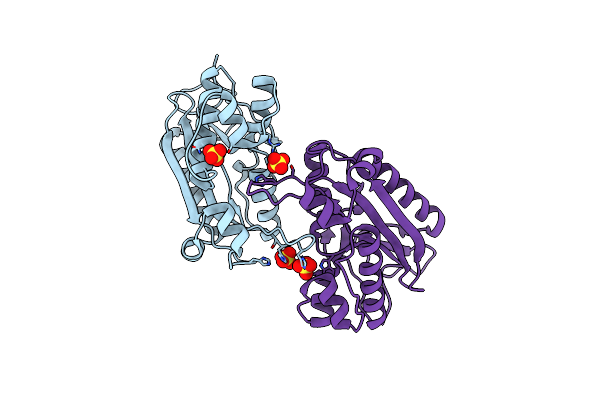 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-07-06 Classification: HYDROLASE Ligands: SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2022-07-06 Classification: HYDROLASE Ligands: AMP, SO4, MN, EDO |
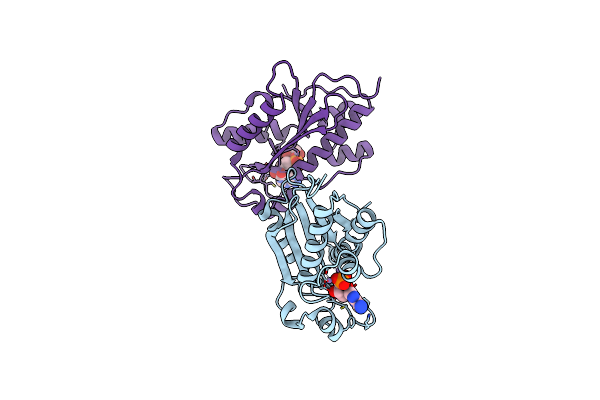 |
Crystal Structures And Ribonuclease Activity Of The Flavivirus Host Factor Eri3 That Is Involved In Viral Rna Synthesis Define The Eri Subfamily Of Structure-Specific 3-Prime - 5-Prime Exoribonucleases
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2022-02-16 Classification: HYDROLASE Ligands: AMP, MN, GOL |
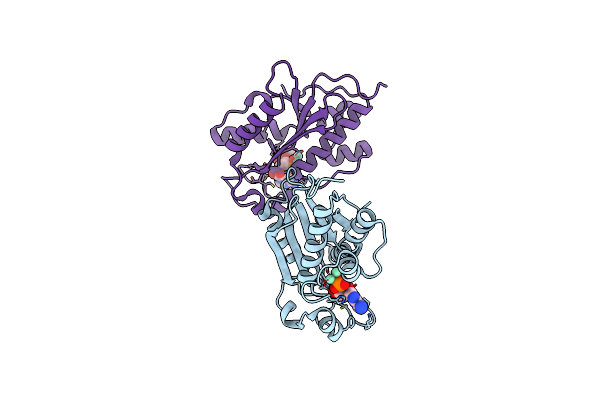 |
Crystal Structures And Ribonuclease Activity Of The Flavivirus Host Factor Eri3 That Is Involved In Viral Rna Synthesis Define The Eri Subfamily Of Structure-Specific 3-Prime - 5-Prime Exoribonucleases
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2022-02-16 Classification: HYDROLASE Ligands: AMP, SM, GOL |
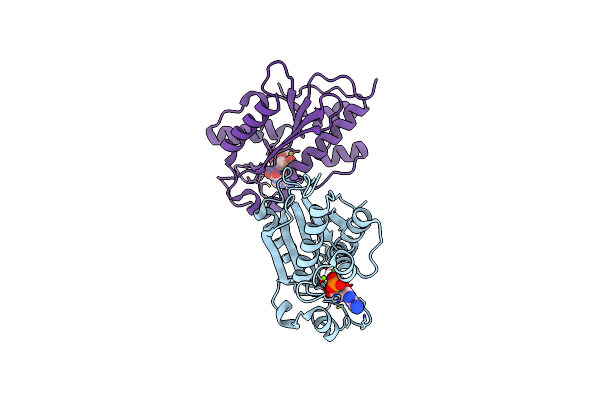 |
Crystal Structures And Ribonuclease Activity Of The Flavivirus Host Factor Eri3 That Is Involved In Viral Rna Synthesis Define The Eri Subfamily Of Structure-Specific 3-Prime - 5-Prime Exoribonucleases
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-02-16 Classification: HYDROLASE Ligands: AMP, MG, GOL |

