Search Count: 104
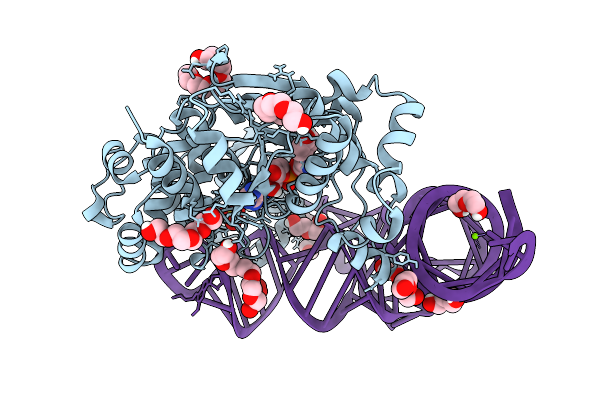 |
Structure At 1.9 A Resolution Of Thermus Thermophilus Tyrosyl-Trna Synthetase Bound To Wild-Type Modified Trna(Tyr), Tyrosine And Amp
Organism: Thermus thermophilus hb27
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: RNA BINDING PROTEIN Ligands: AMP, TYR, PEG, SO4, 1PE, PGE, MG |
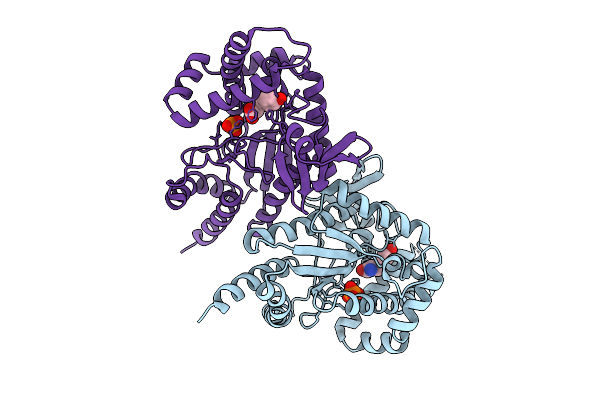 |
Organism: Photorhabdus luminescens
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: TRANSFERASE Ligands: PO4, TYR |
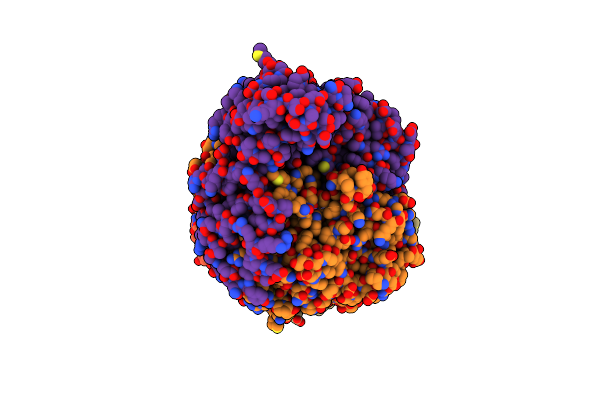 |
Crystal Structure Of Tyrosine Decarboxylase H203F Mutant In Complex With The Cofactor Plp And Tyrosine
Organism: Papaver somniferum
Method: X-RAY DIFFRACTION Resolution:3.35 Å Release Date: 2025-02-12 Classification: LYASE Ligands: TYR |
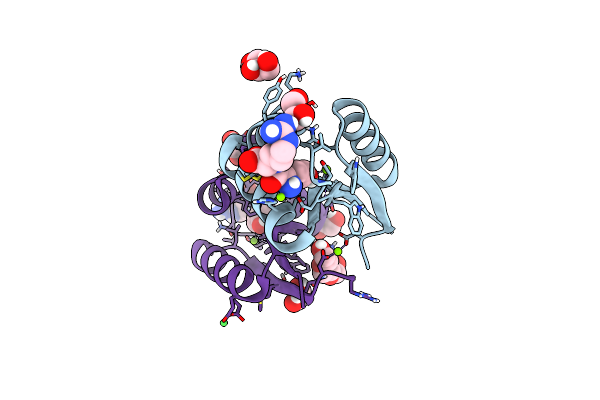 |
Organism: Mycolicibacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:1.28 Å Release Date: 2024-12-11 Classification: PROTEIN BINDING Ligands: TYR, ARG, MG, GOL, EDO, PGE, NI |
 |
Organism: Aetokthonos hydrillicola thurmond2011
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2024-10-02 Classification: METAL BINDING PROTEIN Ligands: FE2, TYR, NI |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-07-10 Classification: MEMBRANE PROTEIN Ligands: TYR |
 |
Small Dipeptide Analogues Developed By Co-Crystal Structure Of Stenotrophomonas Maltophilia Dipeptidyl Peptidase 7
Organism: Stenotrophomonas maltophilia (strain r551-3)
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2023-09-06 Classification: HYDROLASE Ligands: ALC, TYR |
 |
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2022-12-14 Classification: ELECTRON TRANSPORT Ligands: PEE, CDL, 8Q1, PLX, TYR, UQ, ADP |
 |
Crystal Structure Of Mfng, An L- And D-Tyrosine O-Methyltransferase From The Marformycin Biosynthesis Pathway Of Streptomyces Drozdowiczii, With Sah And L-Tyrosine Bound At 1.4 A Resolution (P212121 - Form Ii)
Organism: Streptomyces drozdowiczii
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2022-09-28 Classification: TRANSFERASE Ligands: SAH, TYR, UNL |
 |
Crystal Structure Of Dmats1 Prenyltransferase In Complex With L-Tyr And Dmspp
Organism: Fusarium fujikuroi
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2022-09-28 Classification: TRANSFERASE Ligands: TYR, DST |
 |
Crystal Structure Of Human 14-3-3 Protein Beta In Complex With Cftr Peptide Ps753Ps768 And Ppi Stabilizer Cy007424
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2022-05-25 Classification: PROTEIN TRANSPORT Ligands: ARG, Q95, TYR, GLN |
 |
Crystal Structure Of The L-Tyrosine-Bound Radical Sam Tyrosine Lyase Thih (2-Iminoacetate Synthase) From Thermosinus Carboxydivorans
Organism: Thermosinus carboxydivorans nor1
Method: X-RAY DIFFRACTION Resolution:1.27 Å Release Date: 2022-05-11 Classification: LYASE Ligands: SF4, 5AD, MET, TYR, SO4, K, BR, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2021-12-08 Classification: HYDROLASE/INHIBITOR Ligands: TYR, QOV, GOL |
 |
Organism: Stenotrophomonas maltophilia (strain r551-3)
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2021-11-03 Classification: HYDROLASE Ligands: VAL, TYR |
 |
Organism: Stenotrophomonas maltophilia (strain r551-3)
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2021-11-03 Classification: HYDROLASE Ligands: TYR, GOL |
 |
Organism: Stenotrophomonas maltophilia (strain r551-3)
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2021-11-03 Classification: HYDROLASE Ligands: ASN, TYR, GOL |
 |
Organism: Stenotrophomonas maltophilia (strain r551-3)
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2021-11-03 Classification: HYDROLASE Ligands: PHE, TYR, GOL |
 |
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (L372S, L536S) In Complex With Dmeri-29
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2021-09-08 Classification: TRANSCRIPTION/TRANSCRIPTION INHIBITOR Ligands: 7OR, TYR, SER, CYS |
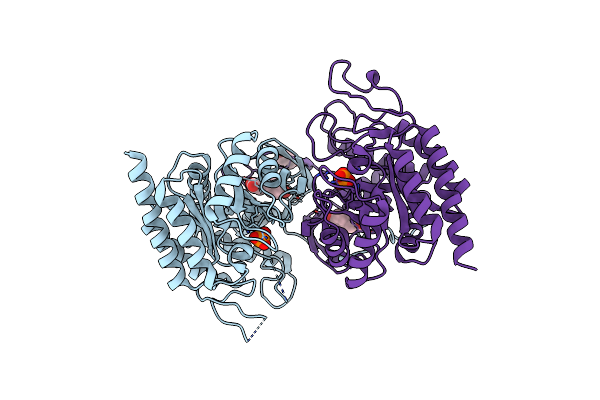 |
Crystal Structure Of Aro4P, 3-Deoxy-D-Arabino-Heptulosonate-7-Phosphate (Dahp) Synthase From Candida Auris, L-Tyr Complex
Organism: Candida auris
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2021-07-28 Classification: LYASE Ligands: TYR, PO4, GOL, CL |
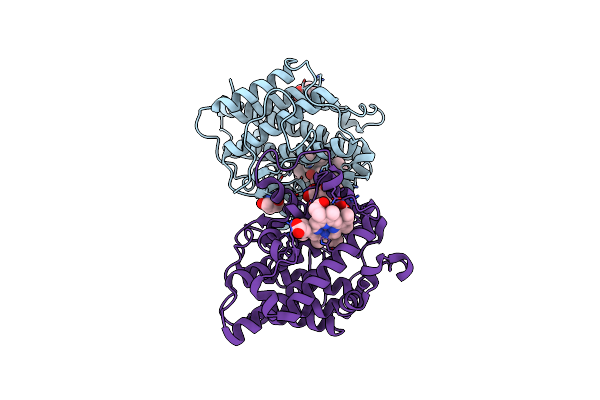 |
A 1.89-A Resolution Substrate-Bound Crystal Structure Of Heme-Dependent Tyrosine Hydroxylase From S. Sclerotialus
Organism: Streptomyces sclerotialus
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2021-03-31 Classification: OXIDOREDUCTASE Ligands: HEM, TYR, BTB, TRS |

