Search Count: 28
 |
N-Terminal Domain Of Restriction Endonuclease Eco15I In The Absence Of Dna.
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.66 Å Release Date: 2024-07-17 Classification: HYDROLASE |
 |
N-Terminal Domain Of Restriction Endonuclease Eco15I With Tetra-Methylated Target Dna.
Organism: Escherichia coli, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2024-07-17 Classification: HYDROLASE Ligands: CA |
 |
Organism: Streptomyces griseus
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2022-08-24 Classification: DNA BINDING PROTEIN Ligands: CA |
 |
Organism: Enterobacteria phage lambda, Synthetic
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2014-11-19 Classification: hydrolase/dna Ligands: CA, PO4 |
 |
Organism: Deinococcus radiophilus
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2014-05-14 Classification: HYDROLASE |
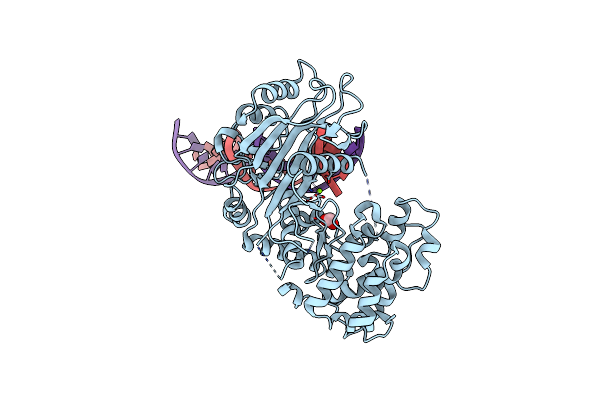 |
Organism: Human herpesvirus 8 type m
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2011-09-21 Classification: DNA BINDING PROTEIN/DNA Ligands: MG, FMT |
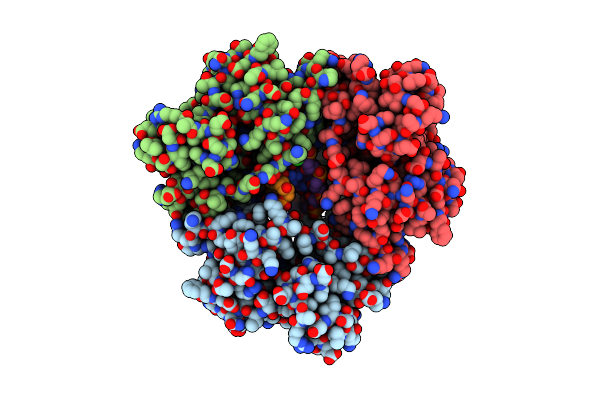 |
Crystal Structure Of Lambda Exonuclease In Complex With A 12 Bp Symmetric Dna Duplex
Organism: Enterobacteria phage lambda
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2011-07-20 Classification: HYDROLASE/DNA Ligands: CA, PO4, CL |
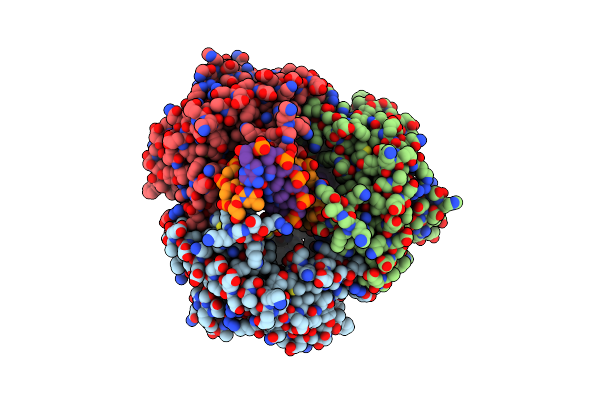 |
Crystal Structure Of The K131A Mutant Of Lambda Exonuclease In Complex With A 5'-Phosphorylated 14-Mer/12-Mer Duplex And Magnesium
Organism: Enterobacteria phage lambda
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2011-07-20 Classification: HYDROLASE/DNA Ligands: PO4, CL, MG |
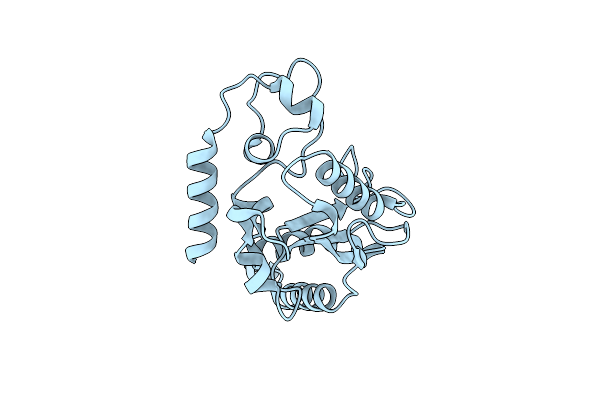 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2010-09-08 Classification: HYDROLASE |
 |
Dna Binding And Cleavage By The Giy-Yig Endonuclease R.Eco29Ki Inactive Variant E142Q
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2010-09-08 Classification: HYDROLASE/DNA |
 |
Dna Binding And Cleavage By The Giy-Yig Endonuclease R.Eco29Ki Inactive Variant Y49F
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2010-09-08 Classification: HYDROLASE/DNA Ligands: PO4 |
 |
Crystal Structures Of Holliday Junction Resolvases From Archaeoglobus Fulgidus Bound To Dna Substrate
Organism: Archaeoglobus fulgidus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2009-05-26 Classification: HYDROLASE/DNA Ligands: HEZ, GOL |
 |
Crystal Structures Of Holliday Junction Resolvases From Archaeoglobus Fulgidus Bound To Dna Substrate
Organism: Archaeoglobus fulgidus dsm 4304, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2009-05-26 Classification: HYDROLASE/DNA |
 |
Crystal Structures Of Holliday Junction Resolvases From Archaeoglobus Fulgidus Bound To Dna Substrate
Organism: Archaeoglobus fulgidus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2009-05-26 Classification: HYDROLASE/DNA Ligands: NCO |
 |
Crystal Structure Of The Beta-Beta-Alpha-Me Type Ii Restriction Endonuclease Hpy99I In The Absence Of Edta
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2009-04-28 Classification: HYDROLASE/DNA Ligands: ZN, NA, 1PE |
 |
Crystal Structure Of The Beta-Beta-Alpha-Me Type Ii Restriction Endonuclease Hpy99I
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2009-03-31 Classification: HYDROLASE/DNA Ligands: ZN, NA, PG4 |
 |
1.6A Resolution Structure Of Archaeoglobus Fulgidus Hjc, A Holliday Junction Resolvase From An Archaeal Hyperthermophile
Organism: Archaeoglobus fulgidus
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2009-03-24 Classification: HYDROLASE Ligands: ACT, NH4 |
 |
1.6A Resolution Structure Of Archaeoglobus Fulgidus Hjc, A Holliday Junction Resolvase From An Archaeal Hyperthermophile
Organism: Archaeoglobus fulgidus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2009-03-24 Classification: HYDROLASE Ligands: CL |
 |
Structure Of The A138T Promiscuous Mutant Of The Ecori Restriction Endonuclease Bound To Its Cognate Recognition Site.
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2007-10-23 Classification: hydrolase/DNA |
 |
Alteration Of Sequence Specificity Of The Type Ii Restriction Endonuclease Hincii Through An Indirect Readout Mechanism
Organism: Haemophilus influenzae
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2006-07-18 Classification: HYDROLASE/DNA |

