Search Count: 21
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.74 Å Release Date: 2016-11-16 Classification: Viral Protein/Inhibitor |
 |
Organism: Homo sapiens, Unidentified
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-11-16 Classification: Viral Protein/Inhibitor |
 |
Crystal Structure Of A Computationally Designed Inhibitor Of An Epstein-Barr Viral Bcl-2 Protein
Organism: Epstein-barr virus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-07-09 Classification: Viral Protein/Inhibitor Ligands: EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2013-03-06 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 1FV |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2013-03-06 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: BX7, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.34 Å Release Date: 2013-03-06 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: BX7, HG, CL |
 |
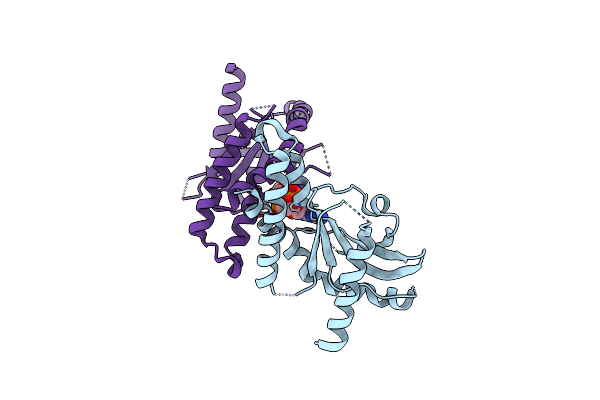
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2012-07-25 Classification: PROTEIN BINDING |
 |
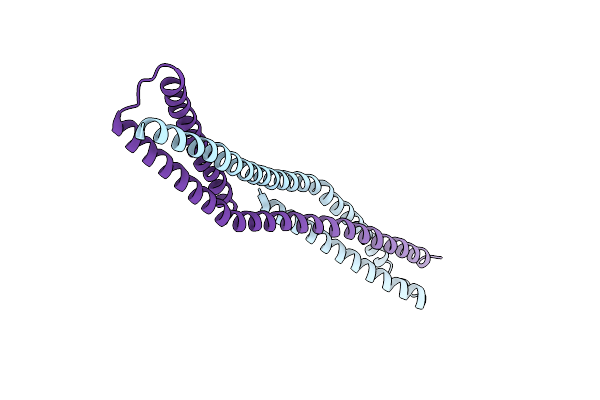
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2012-07-25 Classification: PROTEIN BINDING Ligands: C2E |
 |
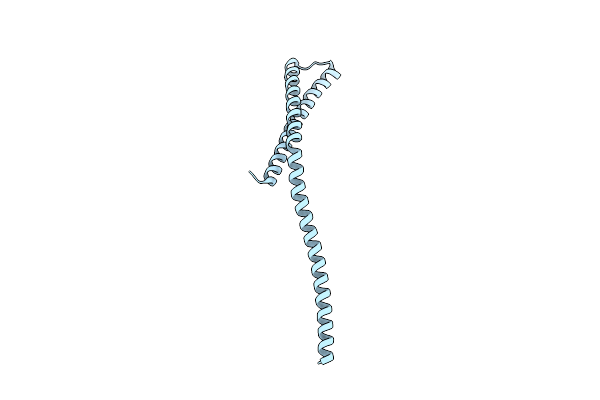
Crystal Structure Of A Domain Of A Protein Involved In Formation Of Actin Cytoskeleton
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2011-10-12 Classification: PROTEIN BINDING |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2011-09-14 Classification: PROTEIN BINDING |
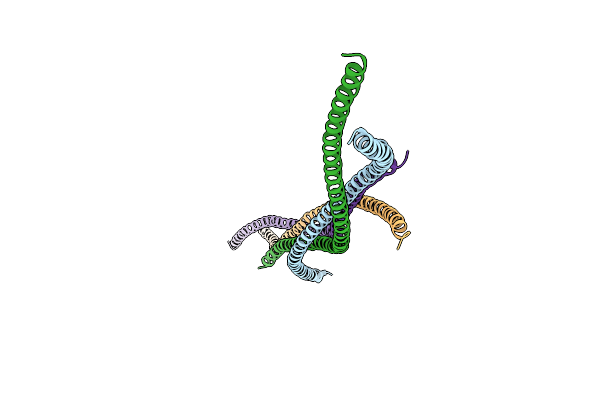 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2011-03-23 Classification: TRANSFERASE |
 |
Crystal Structure Of The Ubiquitin Conjugating Enzyme Ube2G2 Bound To The G2Br Domain Of Ubiquitin Ligase Gp78
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.76 Å Release Date: 2009-02-10 Classification: LIGASE |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.56 Å Release Date: 2007-09-18 Classification: LIGASE Ligands: K |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2007-09-18 Classification: LIGASE |
 |
Crystal Structure Of Azithromycin Bound To The G2099A Mutant 50S Ribosomal Subunit Of Haloarcula Marismortui
Organism: Haloarcula marismortui
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2005-04-26 Classification: RIBOSOME Ligands: ZIT, MG, K, NA, CL, SR, CD |
 |
Crystal Structure Of Erythromycin Bound To The G2099A Mutant 50S Ribosomal Subunit Of Haloarcula Marismortui
Organism: Haloarcula marismortui
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2005-04-26 Classification: RIBOSOME Ligands: ERY, MG, K, NA, CL, CD |
 |
Crystal Structure Of Telithromycin Bound To The G2099A Mutant 50S Ribosomal Subunit Of Haloarcula Marismortui
Organism: Haloarcula marismortui
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2005-04-26 Classification: RIBOSOME Ligands: MG, K, NA, CL, TEL, CD |
 |
Crystal Structure Of Virginiamycin M And S Bound To The 50S Ribosomal Subunit Of Haloarcula Marismortui
Organism: Haloarcula marismortui, Streptomyces virginiae
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2005-04-26 Classification: RIBOSOME/ANTIBIOTIC Ligands: MG, K, NA, CL, VIR, CD |
 |
Crystal Structure Of The Mutant 50S Ribosomal Subunit Of Haloarcula Marismortui Containing A Three Residue Deletion In L22
Organism: Haloarcula marismortui
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2005-04-26 Classification: RIBOSOME Ligands: MG, K, NA, CL, CD |
 |
Crystal Structure Of Clindamycin Bound To The G2099A Mutant 50S Ribosomal Subunit Of Haloarcula Marismortui
Organism: Haloarcula marismortui
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2005-04-26 Classification: RIBOSOME Ligands: CLY, MG, K, NA, CL, CD |

