Search Count: 13
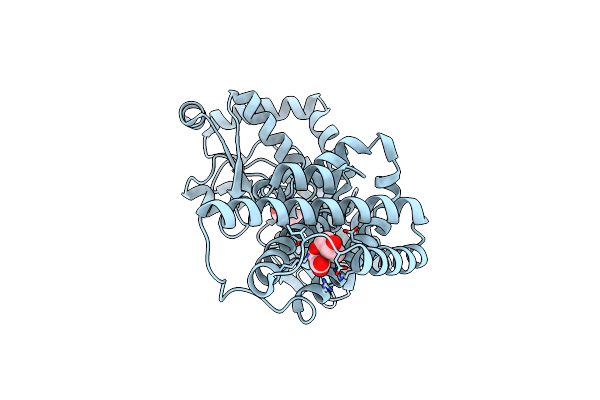 |
Organism: Janthinobacterium sp. hh01
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-07-10 Classification: UNKNOWN FUNCTION Ligands: TSA, PPY, SO4 |
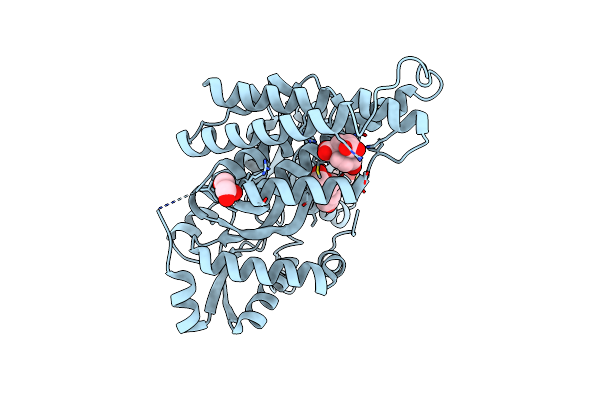 |
Organism: Aequoribacter fuscus
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2024-07-10 Classification: UNKNOWN FUNCTION Ligands: TSA, PEG, GOL, CL |
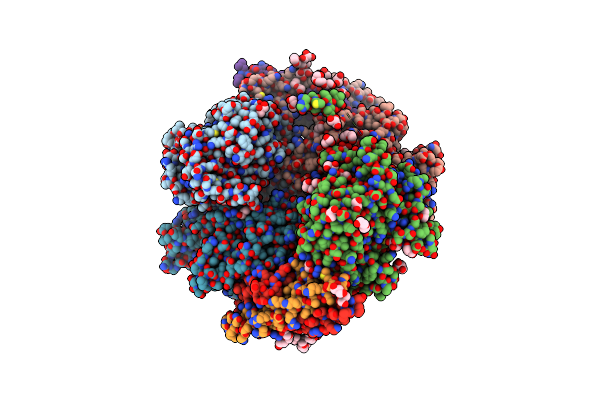 |
Non-Covalent Complex Of And Dahp Synthase And Chorismate Mutase From Corynebacterium Glutamicum With Bound Transition State Analog
Organism: Corynebacterium glutamicum
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2017-08-02 Classification: TRANSFERASE/ISOMERASE Ligands: GOL, PEG, PGE, MN, PO4, TRP, PG4, TSA |
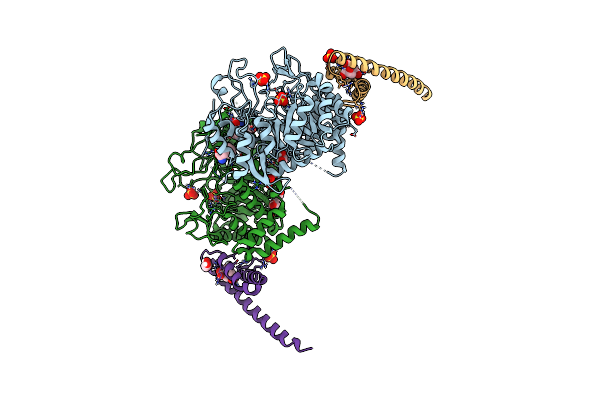 |
Non-Covalent Complex Of Dahp Synthase And Chorismate Mutase From Mycobacterium Tuberculosis With Bound Transition State Analog And Feedback Effectors Tyrosine And Phenylalanine
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2016-03-16 Classification: transferase/isomerase Ligands: MN, GOL, CL, SO4, PHE, TYR, TSA |
 |
Arg90Cit Chorismate Mutase Of Bacillus Subtilis In Complex With A Transition State Analog
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-04-16 Classification: ISOMERASE Ligands: TSA |
 |
Non-Covalent Complex Between Dahp Synthase And Chorismate Mutase From Mycobacterium Tuberculosis With Bound Tsa
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2009-07-07 Classification: TRANSFERASE/ISOMERASE Ligands: MN, SO4, CE1, PG4, GOL, TSA |
 |
Organism: Methanocaldococcus jannaschii
Method: SOLUTION NMR Release Date: 2006-10-31 Classification: ISOMERASE Ligands: TSA |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2006-03-28 Classification: ISOMERASE Ligands: TSA |
 |
Structure Of Yeast Chorismate Mutase With Bound Trp And An Endooxabicyclic Inhibitor
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 1998-01-14 Classification: COMPLEX (ISOMERASE/PEPTIDE) Ligands: TRP, TSA |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 1998-01-14 Classification: COMPLEX (ISOMERASE/PEPTIDE) Ligands: TYR, TSA |
 |
Atomic Structure Of The Buried Catalytic Pocket Of Escherichia Coli Chorismate Mutase
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 1995-12-01 Classification: CHORISMATE MUTASE Ligands: TSA |
 |
Crystal Structures Of The Monofunctional Chorismate Mutase From Bacillus Subtilis And Its Complex With A Transition State Analog
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 1994-07-31 Classification: ISOMERASE Ligands: TSA |
 |
Routes To Catalysis: Structure Of A Catalytic Antibody And Comparison With Its Natural Counterpart
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 1994-05-31 Classification: IMMUNOGLOBULIN Ligands: TSA |

